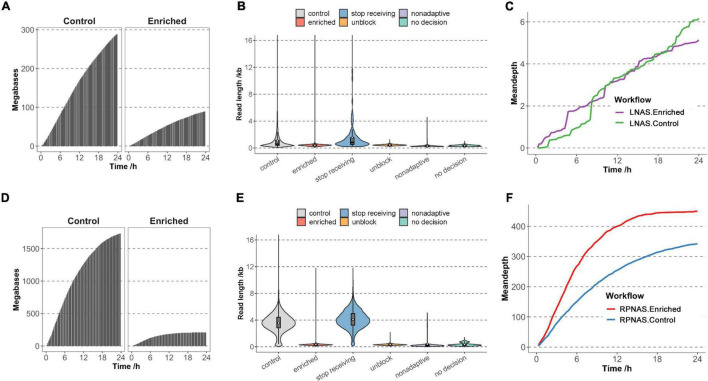
FIGURE 1.
Adaptive sequencing performance of the ligation-based nanopore adaptive sequencing (LNAS) and rapid PCR-based nanopore adaptive sequencing (RPNAS) workflows. The cumulative total base yield in the enriched and control groups over 24 h sequencing of sample M9 with the LNAS workflow (A) and sample HB204 with the RPNAS workflow (D). The read length distribution of sample M9 with the LNAS workflow (B) and sample HB204 with the RPNAS workflow (E). Reads in both the enriched and control groups were represented with red and gray colors, separately. Blue, orange, purple, and green colors represent stop receiving, unblock, non-adaptive, and no decision, respectively. The white dot represents the mean read length of each decision. The mean depth of human adenovirus (HAdV) in both the enriched and control groups over 24 h sequencing of sample M9 with LNAS workflow (C) and sample HB204 with RPNAS workflow (F).