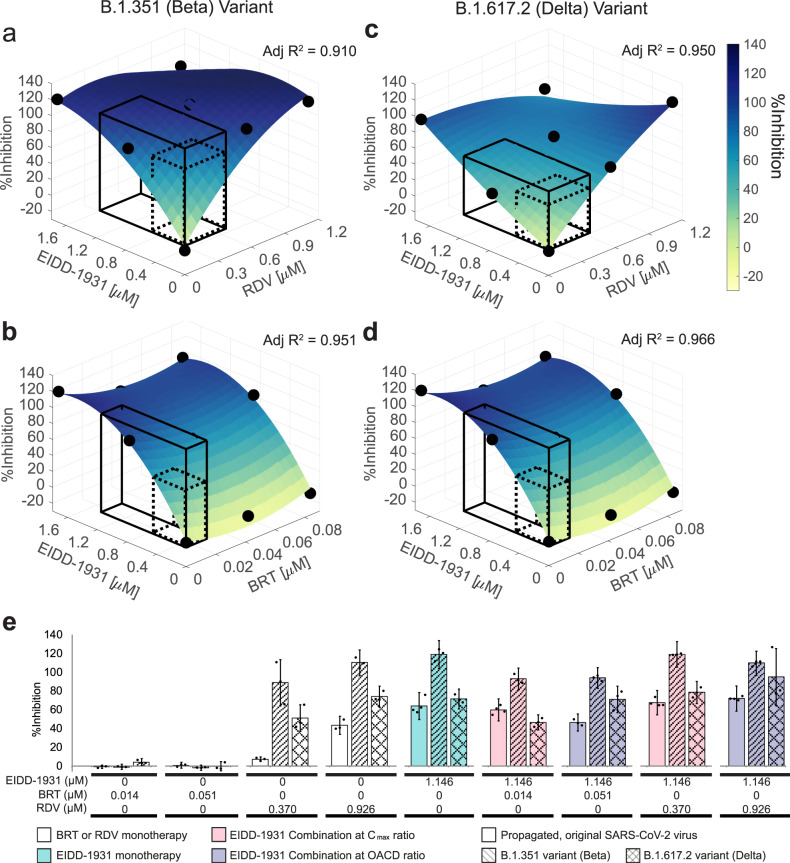
Fig. 4. Validation of EIDD-1931 drug interactions affecting %Inhibition in SARS-CoV-2 B.1.351 and B.1.617.2 variants.
a–d Surface plots of EIDD-1931 interactions with remdesivir (RDV) and baricitinib (BRT) in the validation interaction space, clinically actionable interaction space (black, solid line border) and the interaction space from the IDentif.AI-x analysis (black, dotted line border). All experiments were performed with N = 3 replicates, which were independently included in the surface construction. Black, round markers indicate an average %Inhibition of the replicates for each treatment. Adjusted R2 (Adj R2) indicates goodness of the fit for each interaction surface. The experiments with SARS-CoV-2 B.1.351 variant (a, b), and B.1.617.2 variant (c, d) were performed in two independent sets. e %Inhibition against the propagated, original SARS-CoV-2 strain (bars with block filling), B.1.351 variant (bars with line filling) and B.1.617.2 variant (bars with cross lines filling) of 10% Cmax EIDD-1931 in monotherapy (green) and in a combination with RDV and BRT at two concentration ratios: the ratio dictated by the Cmax values of the drugs (Cmax ratio; pink) and the ratio tested in the IDentif.AI-x experimental set (OACD ratio; purple). Black markers indicate individual data points. Error bars represent propagated standard deviation (s.d). Of note, this propagated s.d. did not arise from the replicates’ spread, but from plate-to-plate variation (s.d. of the controls). No statistically significant difference was detected with Kruskal–Wallis test when followed by Dunn’s post hoc test.