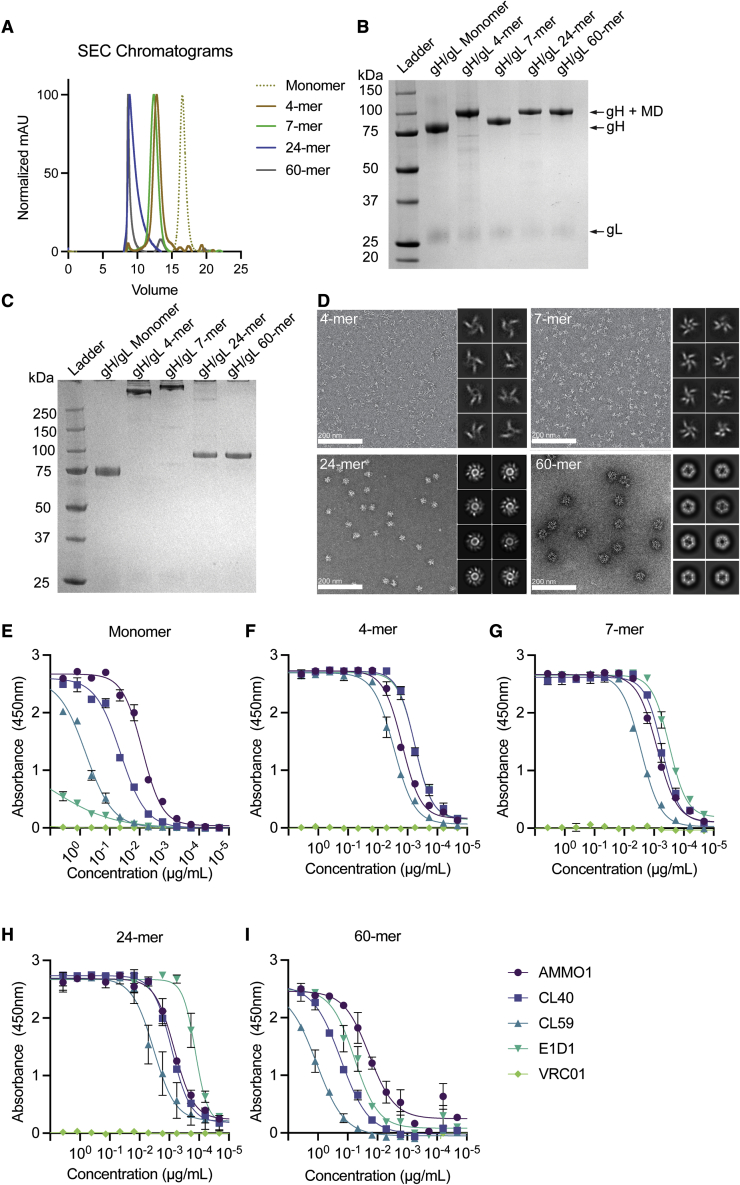
Figure 1.
Biochemical and biophysical characterization of multimeric gH/gL nanoparticles
(A) Monomeric gH/gL and multimeric gH/gL nanoparticles were analyzed by size-exclusion chromatography (SEC) on a Superose 6 column as indicated.
(B) Reducing SDS-PAGE analysis of 1 μg of monomeric gH/gL or multimeric gH/gL nanoparticles. Bands corresponding to gL, gH, and gH fused to 4-, 7-, 24-, or 60-mer multimerization domains (MDs) are indicated with arrows.
(C) Non-reducing SDS-PAGE analysis of 1 μg of the proteins in (B).
(D) Negative-stain electron microscopy was performed on 4-, 7-, 24-, or 60-mer gH/gL nanoparticles as indicated. The eight most frequent 2D class averages for each particle are shown in the inlay. Scale bars represent 200 nm.
(E–I) Binding of the anti-gH/gL mAbs E1D1, CL40, CL59, and AMMO1 to monomeric gH/gL (E) or multimeric gH/gL nanoparticles (F–I) were measured by ELISA as indicated. Each data point represents the mean, and error bars represent the standard deviation of two technical replicates. The anti-HIV-1 Env mAb VRC01 was used as a control for non-specific binding.