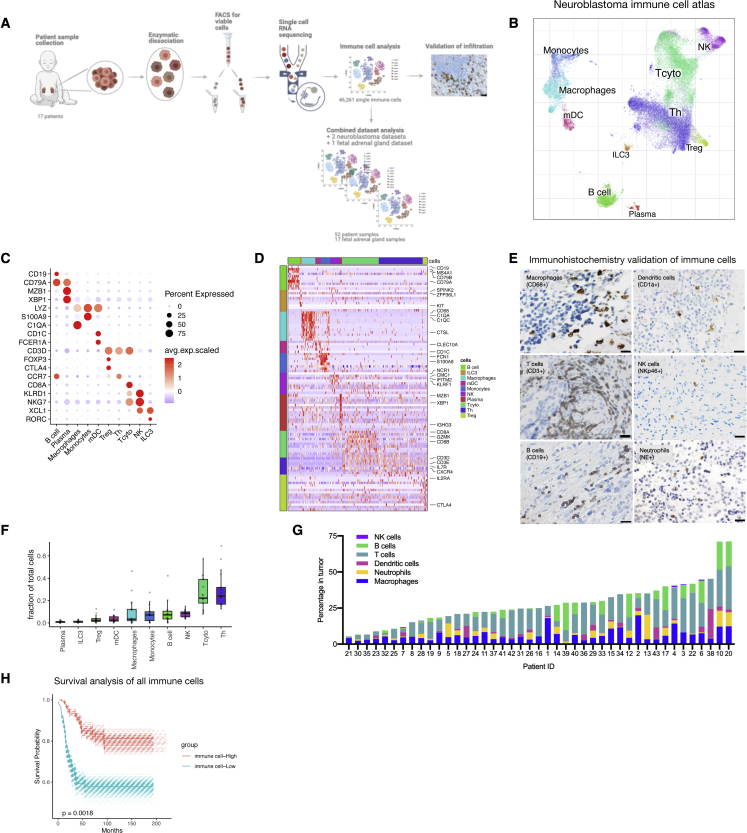
Figure 1.
Global immune cell landscape of human NB
(A) Experimental design: human NB tumor tissue was mechanically and enzymatically dissociated. Immune cells were studied in silico, infiltration validated using immunohistochemistry, and additional data added for combinational analysis.
(B) Global overview of NB immune cell atlas containing 46,261 cells, color coded by annotated cell type (n = 17).
(C and D) (C) Subset marker gene expression and (D) heatmap of marker genes associated with major immune cell types.
(E) Images for macrophages (CD68+), dendritic cells (CD1a+), neutrophils (NE+), NK cells (NKp46+), B cells (CD19+), and T cells (CD3+) in NB tumors. Scale bar, 100 μm. n = 43.
(F) Fraction of cells from all immune cells shown for the different subtypes (n = 17).
(G) Quantification of percentage of cells from (E) (n = 43).
(H) Survival data on bulk RNA-seq data. A gene signature derived from scRNA-seq high was considered the top 25% highest expression of the signature genes, whereas low is the lowest 25% expression of the signature genes (STAR Methods).