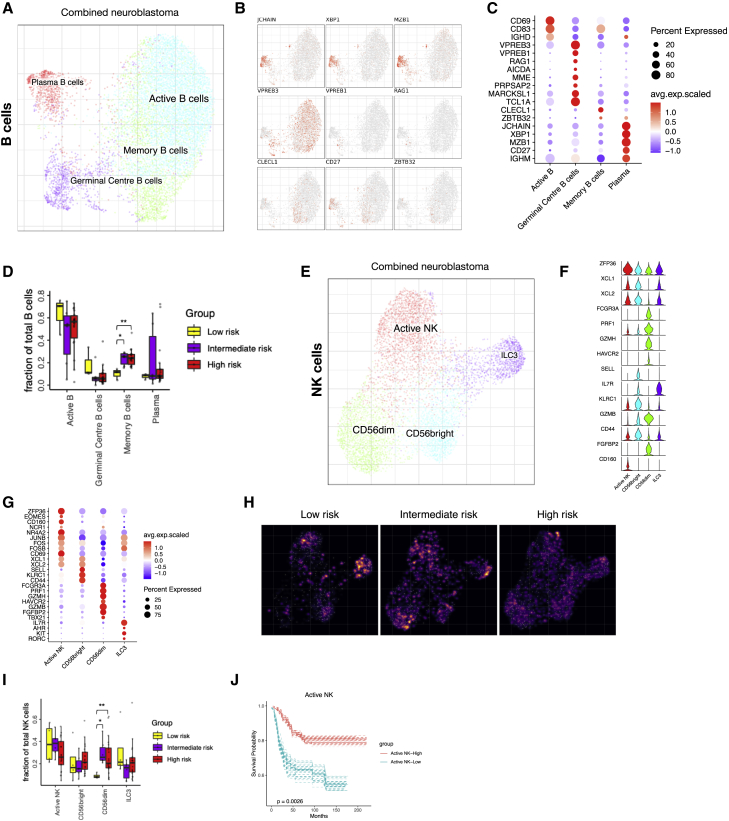
Figure 3.
B and NK cell heterogeneity and infiltration in NB tumors
(A–C) (A) Detailed annotation of the B cells subpopulations is shown on a B cell-specific joint embedding combining multiple NB datasets. Key marker gene expression shown in feature plots (B) and in a dotplot (C) for the different subpopulations of B cells.
(D) The fraction of B cell subtypes comparing low- (n = 3), intermediate- (n = 6), and high-risk (n = 18) NB. Wilcoxon rank-sum test was used for statistical analysis; ∗p < 0.05, ∗∗p < 0.01.
(E–G) (E) Detailed annotation of NK cell subpopulations shown on NK cell-specific embedding from the combined dataset. Key marker gene expression shown in a violin plot (F) and in a dotplot (G) for the different subpopulations of NK cells.
(H) Density plot for NK cell-specific embedding showing the number of cells in low-, intermediate-, and high-risk NB. Brighter color corresponds to a denser region.
(I) The fraction of cells in the different NK cell populations comparing low- (n = 4), intermediate- (n = 6), and high-risk (n = 21) NB. Wilcoxon rank-sum test was used for statistical analysis; ∗p < 0.05, ∗∗p < 0.01.
(J) Survival curve for active NK cells.