Fig. 1.
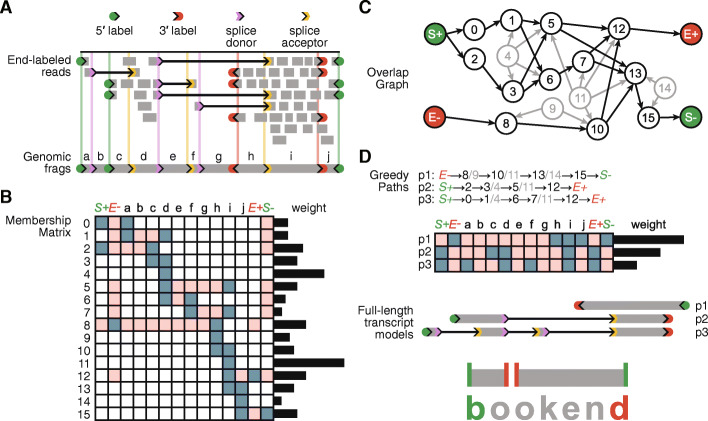
End-guided assembly with Bookend. A Individual RNA-seq reads are mapped to a genome, recording which reads mark a transcript 5′ or 3′ end, and which reads span one or more splice junctions. Ranges between adjacent features are recorded as frags. B Each unique read structure is recorded in a condensed representation as one element in a Membership Matrix; blue—included, pink—excluded. The weight of each element is the coverage depth of matching reads (sequenced bases/length) across the element. C A directed graph is constructed between overlapping elements of the Membership matrix. Weights of contained elements (gray) are distributed proportionally to their containers. D A set of optimal paths through the graph is iteratively constructed from the heaviest unassigned elements. Complete Paths are output as full-length transcript annotations