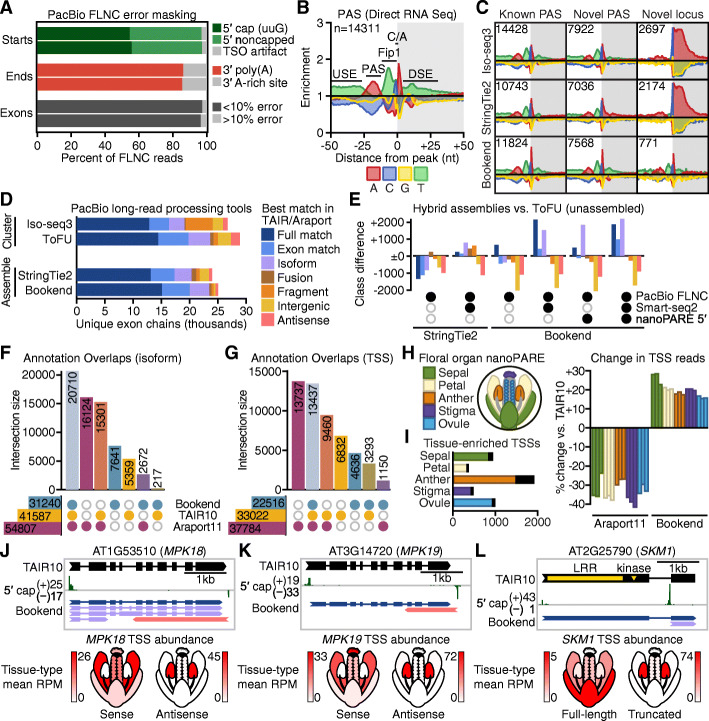
Fig. 3.
Long-read sequencing is augmented by hybrid assembly. A Artifacts identified in PacBio FLNC reads from two SMRT cells by alignment to the Arabidopsis reference genome. B Nucleotide frequency enrichment in a ± 50 nt window around poly(A) sites (PAS) identified by Direct RNA-seq [30]. C Nucleotide enrichment around 3′ ends of transcripts constructed from PacBio reads by Iso-seq3 (top), StringTie2 (middle), and Bookend (bottom) at sites overlapping a TAIR10 PAS (left), novel PAS at a known gene (middle), and novel antisense or intergenic loci (right); colors and scales as in (B). D Classification against the closest match in TAIR10 or Araport11 of transcripts constructed by four long-read processing strategies: Iso-seq3 clustering, cluster collapse by ToFU, and FLNC assembly by StringTie2 or Bookend. E Effect of long-read assembly on the number of transcripts by class (colored as in D) by StringTie2 (left) or Bookend (right) using hybrid assembly with one or more tissue-matched sequencing libraries. Bars show difference vs. ToFU-collapsed Iso-seq3 clusters. F UpSet plot depicting the number of overlapping transcript isoforms present in TAIR10, Araport11, and the Bookend Floral Bud assembly. G UpSet plot for the union of transcription start site peaks, allowing a ± 50 nt overlap window. H Diagram of floral organs analyzed by nanoPARE (left) and percent change in the number of TSS-overlapping reads for alternative annotations to TAIR10 in 15 tissue-specific nanoPARE libraries (right). I Bar chart of the number of enriched TSSs in each tissue type (≥4-fold mean RPM ingroup vs. outgroup, ANOVA p < 0.01, Benjamini-Hochberg correction). Shaded portions are TSSs exclusive to Bookend Floral Bud. J IGV browser image depicting Bookend Floral Bud assemblies for MPK18 (top) and nanoPARE abundance heatmaps for MPK18 sense and antisense TSSs (bottom). K Architecture and abundance of MPK19 isoforms as in (K). L Full-length and truncated isoforms of SKM1 assembled by Bookend (top). Heatmaps as in (J–K) for SKM1 TSSs (bottomt). TPM, transcripts per million; TSS, transcription start site