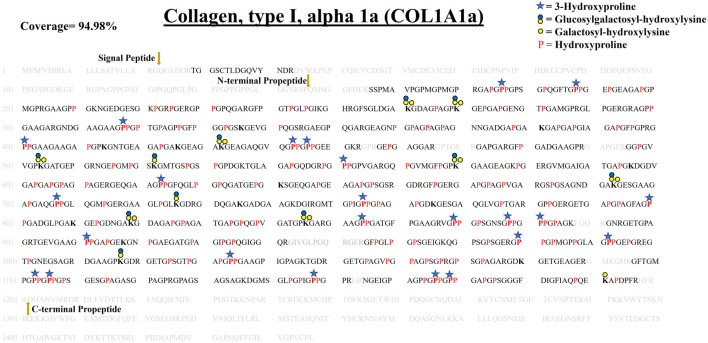
FIGURE 3.
Comprehensive map of proline/lysine hydroxylation sites and lysine O-glycosylation sites in COL1A1a of WT zebrafish heart ECM. Identified peptide sequence in the proteomic analysis is shown in black color, sequence not identified in this analysis are shown in grey color. A total of 94.98% sequence coverage of COL1A1a is detected (considering the matured form of COL1A1The signalgnal peptide is 22 amino acids (1–22) long. Sequence alignment matching with human COL1A1 revealed the propeptide cleavage sites. Dark yellow arrows show N terminal (23–146) and C-Term (1,202–1,447) propeptide cleavage sites. As shown in the top right corner, red bold “P” with a blue star represents 3-hydroxyproline on the Xaa position followed by 4-hydroxyproline the on Yaa position in the Gly-Xaa-Yaa motif. 4-hydroxyproline on Yaa position is represented with red color “P”. Hydroxyproline on unusual Xaa position with (Ala, Val, Met, Ile, Ser, Glu, Arg, and Asp) on Yaa position are also identified but cannot label either 3-hydroxyproline or 4-hydroxyproline. Hydroxylysine sites are presented by bold “K”. Lysine sites highlighted with a yellow circle represents galactosyl-hydroxylysine sites and yellow plus blue coloued circles represent glucosylgalactosyl-hydroxylysine sites. The presence of glucosylgalactosyl-hydroxylysine, galactosyl-hydroxylysine, and hydroxylysine on the same site shows lysine microheterogeneity. A summary of these site-specific PTMs of COL1A1a is presented in Table 1, and all the PSMs for O-glycosylated lysine and 3-hydroxyproline sites are provided in Supplementary Figure S2.1-S2.21.