Significance
Advances in DNA sequencing have provided an unprecedented view of the complex microbial communities that populate global ecosystems. We present a metatranscriptomic analysis of samples from the boreal forest—the largest terrestrial carbon store—capturing the seasonally resolved transcriptomes of Norway spruce roots and more than 350 root-associated fungal species. Our findings link the functional response of host-trees to increased nutrient availability, with profound perturbations in the fungal community. Notably, we observed an exchange in prevalence and host-coordination of specialist ectomycorrhizal species critical for enzymatic cycling of recalcitrant carbon, to metabolically versatile species with resilient melanized cell walls. This research unites kingdom-spanning taxonomic and functional details of the boreal root microbiome, contributing a missing perspective toward modeling global carbon cycling.
Keywords: metatranscriptome, host–microbe, ectomycorrhiza, carbon storage, fungal effectors
Abstract
Carbon storage and cycling in boreal forests—the largest terrestrial carbon store—is moderated by complex interactions between trees and soil microorganisms. However, existing methods limit our ability to predict how changes in environmental conditions will alter these associations and the essential ecosystem services they provide. To address this, we developed a metatranscriptomic approach to analyze the impact of nutrient enrichment on Norway spruce fine roots and the community structure, function, and tree–microbe coordination of over 350 root-associated fungal species. In response to altered nutrient status, host trees redefined their relationship with the fungal community by reducing sugar efflux carriers and enhancing defense processes. This resulted in a profound restructuring of the fungal community and a collapse in functional coordination between the tree and the dominant Basidiomycete species, and an increase in functional coordination with versatile Ascomycete species. As such, there was a functional shift in community dominance from Basidiomycetes species, with important roles in enzymatically cycling recalcitrant carbon, to Ascomycete species that have melanized cell walls that are highly resistant to degradation. These changes were accompanied by prominent shifts in transcriptional coordination between over 60 predicted fungal effectors, with more than 5,000 Norway spruce transcripts, providing mechanistic insight into the complex molecular dialogue coordinating host trees and their fungal partners. The host–microbe dynamics captured by this study functionally inform how these complex and sensitive biological relationships may mediate the carbon storage potential of boreal soils under changing nutrient conditions.
Complex microbial communities (microbiomes) are intrinsically associated with all higher organisms and can contribute functions spanning a continuum, from essential mutualistic services to parasitism (1). DNA-sequencing approaches have greatly increased our understanding of the complexity of microbiomes, from environments as diverse as the human gut (2), to oceans (3) and soils (4). Recent sequencing approaches to measure gene expression within microbial communities (metatranscriptomics) have enabled researchers to complement these taxonomic surveys with real-time functional insights in a range of clinical (5, 6) and ecological (7, 8) settings. Metatranscriptomics can also capture the host transcriptome, providing insight into both community functions and their interaction with active host processes (9, 10). To date, the technical difficulties associated with metatranscriptomics have largely limited its application to controlled laboratory experiments or to interactions between a host and select symbiotic partners. These experiments are essential to generate detailed molecular-level mechanistic understanding of the interaction between single microbial species and their host, but fail to provide understanding of how those species will interact within the complex communities present in nature.
Soil microbes, such as fungi, are primary mediators of carbon (C) and nitrogen (N) cycling in terrestrial ecosystems, and thus have a critical influence on nutrient availability and soil C sequestration and storage (11, 12). In addition, fungi directly interact with over 85% of all vascular plant species via mycorrhizal symbiosis (13), a form of mutualism that has shaped the evolution and expansion of terrestrial plant life. The success of this symbiotic association has resulted in the frequent adoption of mycorrhizal strategies throughout fungal evolution, with current estimates suggesting 40,000 to 50,000 fungal species can form mycorrhiza with plant hosts (14). In doing so, these fungi have experienced varying degrees of diminished autonomy, as they have successively lost biological functions connected to their pathogenic or saprotrophic origins (15). For these kingdom-spanning associations to establish, a complex molecular dialogue must occur to structurally and physiologically coordinate the host with its prospective fungal partners. A crucial component of these interactions are microbial effectors: small secreted proteins deployed by microbes (both mutualists and pathogens) that promote colonization via modulation of host defense mechanisms (16), alteration of cellular architecture (17), or appropriation of host physiology (18).
In the nutrient-limited (NL) soils of boreal forests, ectomycorrhizal (ECM) fungi predominate, and play vital roles in mobilizing mineral nutrients, such as N and phosphorus (P), exchanging them with plant roots for energy-rich C compounds, such as simple sugars (19). Currently, nutrient availability is a critical limitation of boreal plant productivity and C storage, and a restraining force maintaining host-plant dependency on their ECM partners (20). Anthropogenic N deposition and higher rates of mineralization linked to global warming are predicted to diminish this dependency and reduce growth limitations (21), leading to decreases in the proportion of photosynthates allocated belowground and increases in aboveground vegetative C storage, ranging from 5 to 30 parts of C sequestered per unit of additional N (22–24). Until recently, aboveground plant litter was regarded as the primary origin of soil-sequestered C. However, a recent study using bomb-14C modeling showed that at least half of the stored C in boreal soils is derived from plant roots and fungal mycelium (25). As these forests comprise the largest terrestrial biome and harbor more stored C than temperate and tropical forests combined (26, 27), alterations to the diversity, composition, and function of soil- and root-associated microbial communities resulting from increased nutrient availability, particularly N, could be expected to impact ecosystem productivity and stability, with global consequences for C storage and release (28–30).
Our capacity to predict the impact of climate change and anthropogenic influences on a range of ecosystems—and the effect this will have on the global C budget—requires rigorous understanding of the complex microbial communities involved and their interactions with host species. An essential component of this is to understand the diverse biological processes these microbial communities contribute to the global C budget, and how these processes might be altered by changes in symbiotic coordination. Variations in the host–soil microbiome dynamic can be driven by environmental perturbations, either via direct anthropogenic inputs, such as fertilization, or indirect inputs, such as increased atmospheric N deposition or enhanced mineralization rates in warming soils, and can vary along natural nutrient or hydraulic gradients. To begin to develop mechanistic insight into how changes in symbiotic coordination may occur in environments as complex as forest soils, we generated high temporal resolution metatranscriptomes to profile Norway spruce [Picea abies L. (Karst.)] fine roots and over 350 root-associated fungal species over a growing season from NL) and long-term nutrient-enriched (NE) plots in the boreal region of northern Sweden (Materials and Methods and SI Appendix, Supplementary Text and Fig. S1A) and developed robust and reproducible data-analysis pipelines that can be applied broadly to study host–microbiome coordination. This resource is publicly available as the Boreal Rhizospheric Atlas (BRA) online tool to facilitate community utilization (https://www.boreal-atlas.info/). Our findings link the mechanisms by which trees limit C flow and augment defense processes as nutrient availability increases with the profound effect this can have on fungal community structure, function, and host–microbe coordination.
Results
Alterations to Fungal Community Structure.
Field-sampled fine roots of Norway spruce contained RNA originating from the host tree and associated fungal communities. Sequencing this RNA (RNA-seq) revealed a significantly greater relative proportion of fungal RNA-seq reads in samples from NL compared to NE treatment (Fig. 1A) (P < 0.001; Wilcoxon signed-rank test), and supporting microbial phospholipid fatty acids data collected from soils at this site, which showed decreased fungal abundance in NE soils (31). NE conditions had a profound effect on the fungal community transcriptome at all time points throughout the growing season (349 up-regulated and 573 down-regulated differentially expressed [DE] Kyoto Encyclopedia of Genes and Genomes orthologs across the season; adjusted P < 0.01) while there was no apparent signal of seasonality (Fig. 1B). Taxonomic profiling of the data revealed shifts in community structure that were confirmed in the subsequent year using metatranscriptomics and DNA ITS amplicon data (2012) (SI Appendix, Fig. S2).
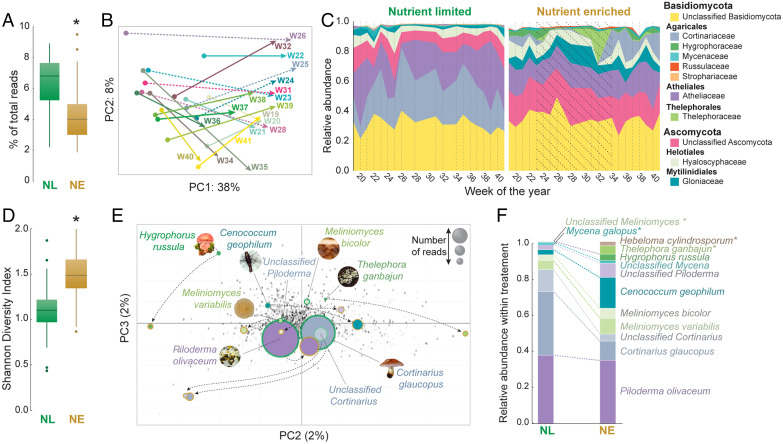
Fig. 1.
Transcriptomic overview of root-associated fungal community structure in samples from NL and NE plots from the same forest site over a growing season. (A) Relative distribution of fungal reads observed per time point in NL and NE conditions. An asterisk (*) indicates significant difference between NL and NE (P < 0.001; two-tailed Wilcoxon signed-rank test). (B) Vector plot of the PCA of a fungal metatranscriptome based on samples collected at 19 time points over a growing season, in NL or NE soils. Each vector denotes the distance and orientation between the average of the NL replicates for a given time point (dot at the origin of the arrow), and those of the NE samples (point of the arrow). Weeks with active fertilization occurring have been indicated by a dotted vector. (C) The number of reads assigned to each fungal family were normalized to the total number of fungal reads present and the top 10 most represented fungal families in each treatment were identified (11 families in total), accounting for 98.7% of all fungal reads. The normalized read count of these 10 families were plotted across the sampling time course for both NL and NE conditions. The period of active fertilization has been indicated by dotted shading. (D) Shannon diversity index of fungal families in NL and NE forest plots. An asterisk (*) indicates significant difference between NL and NE (P < 0.001; two-tailed Wilcoxon signed-rank test). (E) PCA of fungal reads mapped to specific reference genomes (fungal species) in NL and NE conditions. The nine fungal species with the highest read representation are indicated. PC1 was largely attributable to the number of reads assigned to a specific species (86% variance explained) and was visualized using node size. Node color represents species identity and the colored line around each node indicates treatment, with green = NL, gold = NE. (F) Relative hierarchy of the 11 most represented fungal species in NL and NE, representing 49.9% of all fungal reads. An asterisk (*) indicates this species is not in the corresponding list in the opposing treatment.
Briefly, 150 fungal families were identified associated with NL roots, with the most abundant belonging to the phylum Basidiomycota, of which the families Cortinariaceae (27.6%) and Atheliaceae (22.6%) predominated, followed by the Ascomycetes Hyaloscyphaceae (3.4%), and Gloniaceae (1.6%) (Fig. 1C). The majority of reads within each of these families were assigned to a single reference genome: 1) Cortinarius glaucopus (Cortinariaceae), 2) Piloderma olivaceum (Atheliaceae), and 3) Cenococcum geophilum (the only species of the Cenococcum genus; Gloniaceae). For Hyaloscyphaceae, reads were primarily assigned to two reference genomes: Meliniomyces bicolor and Meliniomyces variabilis (synonymy: Hyaloscypha). These taxonomic assignments were conditional to a ≥85% similarity to the available reference genomes (SI Appendix, Supplementary Methods) and may represent a number of closely related species belonging to the same genus. For example, amplicon data (SI Appendix, Fig. S2) identified 10 swarm operational taxonomic units assigned as Piloderma and over 50 as Cortinarius. Despite this caveat, for the remainder of this article the full species names of the best-matching reference genomes are used to avoid confusion.
Fungal family diversity increased significantly in NE conditions (P < 0.001; Wilcoxon rank sum test) (Fig. 1D), accompanied by decreased read representation of formerly dominant Co. glaucopus (from 27.6 to 7.0%) (Fig. 1E). Similarly, NE conditions resulted in a ∼50% reduction in read abundance assigned to P. olivaceum; however, this remained the most dominant species with the reduction reflecting a decrease in overall fungal abundance rather than a complete alteration in fungal abundance hierarchy (Fig. 1 E and F). While reads assigned to these Basidiomycota decreased in NE conditions, read abundance and species dominance increased for the Ascomycota Ce. geophilum (from 1.6 to 6.8%) and Meliniomyces spp. (from 3.4 to 4%) (Fig. 1E).
Remodeling of Fungal Functions.
While conventional amplicon-based approaches assayed community structure (SI Appendix, Fig. S2A), metatranscriptomics revealed the functional impact of community restructuring within a coexpression network. Broadly speaking, the network divided into two hemispheres: 1) genes with higher average transcript abundance in NL conditions and 2) genes with higher average transcript abundance in NE conditions (Fig. 2A and SI Appendix, Fig. S3). Hemisphere 1 was overrepresented in reads assigned to Co. glaucopus or P. olivaceum (P < 0.001; one-proportion z-test), with functional enrichment of cell cycle processes and meiosis, amino acid and secondary metabolite biosynthesis, and signaling pathways (e.g., AMPK, MAPK, TGF-β, ErbB, and calcium signaling pathways; adjusted P < 0.05 in all cases; Fisher exact test) (SI Appendix, Fig. S3). In contrast, hemisphere 2 was overrepresented in reads assigned to Ce. geophilum or Meliniomyces spp., and enriched in functions indicative of carbohydrate starvation in fungal symbionts (32), such as autophagy (e.g., lysosome, glycan degradation, protein processing in the endoplasmic reticulum) and alternate energy pathways (e.g., fatty acid metabolism, glyoxylate cycle, and gluconeogenesis). Functions linked to primary energy metabolism (e.g., pyruvate metabolism, tricarboxylic acid cycle, and oxidative phosphorylation) were also significantly enriched. Taken together, these changes indicate that although exhibiting transcriptional hallmarks of starvation, the metabolic plasticity of species within hemisphere 2 allows them to effectively utilize alternative energy sources (e.g., lipids) and persist in NE soils.
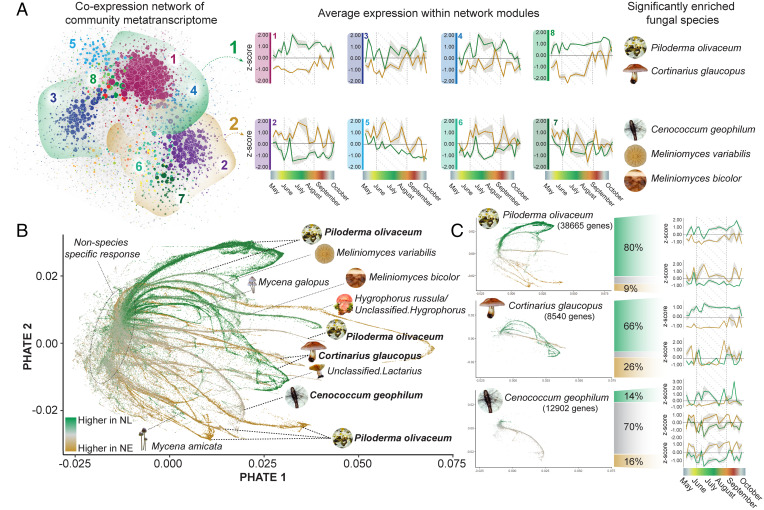
Fig. 2.
Remodeling of fungal community metatranscriptome in samples from NL and NE plots from the same forest site over a growing season. (A) Visualization of the root-associated fungal community network of samples collected from NL and NE forest plots over a growing season. Average expression profiles of the eight largest network modules are displayed, with NL in green and NE in gold, and the SD indicated by gray shading. Significantly enriched functional categories (adjusted P < 0.05; Fisher exact test) for each network module can be found in SI Appendix, Fig. S3. The network modules were grouped into two hemispheres, with hemisphere 1 (indicated in green) displaying higher average expression in NL conditions, and hemisphere 2 (indicated in gold) displaying higher average expression in NE conditions. Fungal species that are significantly enriched in each hemisphere (P < 0.0001; one-proportion z-test) are indicated. (B) Low-dimensional visualization of all fungal genes in both NL and NE conditions. The gene nodes have been colored based on the fold-change differences between NL and NE conditions. Where possible, the branching arms have been assigned to distinct taxonomic assignments, with species names reflecting the reference genome to which RNA-seq reads were assigned. (C) The genes of the three fungal species with the highest assignment of RNA-seq reads (mapped to a specific reference genome) were isolated and separated based on three criteria: 1) fold-change was <−0.5 (higher abundance in NL), indicated in green; 2) fold-change was >0.5 (higher abundance in NE), indicated in gold; and 3) the fold-change was < 0.5 but >−0.5. The average expression profiles of the genes meeting these three criteria have been visualized and corresponding functional enrichment analysis can be found in SI Appendix, Fig. S4.
Taxon-specific expression profiles (Fig. 2 B and C) and associated functional modifications (SI Appendix, Fig. S4) were identified for the three species with the highest representation in the fungal read pool (P. olivaceum, Co. glaucopus, and Ce. geophilum). In response to NE conditions, P. olivaceum and Co. glaucopus underwent extensive transcriptional modulation, with decreased transcript abundance of functions linked to cell growth, nuclear pore complexes, and proteolytic processes, reflecting a decrease of growth in these species. In contrast, the impact of NE conditions on the transcriptome of Ce. geophilum was comparatively subtle, indicating that the observed increase in community dominance did not result from a profoundly altered lifestyle strategy. Functional enrichment analysis of genes that remained stable between treatments (70% of Ce. geophilum genes) revealed a high level of similarity with functions that were prevalent in P. olivaceum during NL but lost in NE. These functions were associated with metabolism and growth: for example, the degradation of polycyclic aromatic hydrocarbons (such as lignin), aromatic amino acid metabolism, and cell growth. Unique to Ce. geophilum in both conditions was the production of bioactive molecules with structural functions, such as melanin biosynthesis (tyrosinase and polyketide synthase); intracellular receptors of fungal innate immunity, which play a role in the detection of nonself and mediating symbiotic cross-talk (signal transducing ATPases containing WD, ANK, and TPR repeat domains) (33); and antimicrobial functions (nonribosomal peptide [NRP] synthetase component F and NRP synthetase/α-amidoadipate reductase, isopenicillin N synthase, and phospholipase A2). This reflects Ce. geophilum navigating interactions with competing microbes (34, 35) and suggests its success under NE conditions is a combination of metabolic versatility, environmental resilience, and chemical suppression of microbial competitors.
Norway Spruce Root Transcriptome.
The temporally resolved Norway spruce root transcriptome revealed extensive seasonal remodeling common to both NL and NE conditions (Fig. 3A) (4,671 up-regulated and 4,999 down-regulated DE genes across the season; Benjamini–Hochberg adjusted P < 0.01; absolute log2 fold-change 0.5). Functional enrichment tests for sets of coexpressed genes within the inferred coexpression network (network modules) were carried out using two complementary ontology-driven approaches: 1) gene ontology (GO) (36), with adjusted P < 0.05; parent–child adjusted significance test (37); and 2) Mapman (38), with adjusted P < 0.05, Fisher exact test. This analysis revealed seasonal transcriptomic alterations primarily indicative of growth, including functions associated with: 1) cell cycle, cell proliferation, and cytoskeleton microtubular networks; and 2) cell wall biosynthesis and remodeling, which coincided with warming soil temperatures, increases in water availability, and a maxima in daylight hours (SI Appendix, Figs. S1 B–D and S5) (network modules 3 and 5). In contrast, the impact of NE had more subtle effects on the root transcriptome than the effect of seasonality (1,689 up-regulated and 956 down-regulated DE genes; NE vs. NL).
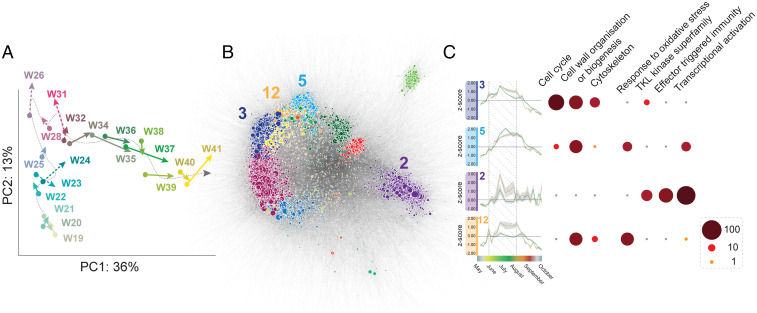
Fig. 3.
Transcriptomic overview of Norway spruce fine roots sampled from NL and NE plots over a growing season. (A) Vector plot of the PCA of Norway spruce fine roots collected at 19 time points over a growing season. Each vector denotes the distance and orientation between the average of the NL replicates for a given time point (dot at the origin of the arrow), and those of the NE samples (point of the arrow). Weeks with active nutrient enrichment are indicated by a dotted vector. (B) Visualization of Norway spruce fine root coexpression network of samples grown on NL and NE forest sites over a growing season. The network has been clustered into distinct network modules and key modules mentioned in the text are indicated. (C) The average expression profiles of the network modules mentioned in the text are indicated for NL (green) and NE (gold), and the SD is indicated by gray shading. A dot plot illustrates significantly (adjusted P < 0.05) enriched GO and MapMan functional categories for these network modules. Average expression profiles and significantly enriched functional categories for all network modules can be found in SI Appendix, Fig. S5.
Broadly speaking, NE conditions did not fundamentally alter seasonal expression, impacting instead the magnitude of the expression dynamics. For example, in both NL and NE samples, genes belonging to network modules 2 and 12 were characterized by a series of transient bursts in transcript abundance across the season, with the most extreme occurring at week 26 (June 27 to July 3). Although temporally coincident, the magnitude of this induction was greater in NE samples (Fig. 3B) (network modules 2 and 12). These modules were significantly enriched in functions associated with signaling and mounting defense responses, such as effector-triggered immunity signaling (nucleotide-binding leucine-rich repeat [NLR] effector receptors and protein kinases), cell wall biogenesis and thickening, oxidative stress, and jasmonic acid biosynthesis. Dehydration-responsive element binding (DREB)-type transcription factors were also significantly enriched in these modules, and overexpression of this class of transcription factor has been linked to enhanced biotic and abiotic stress tolerance in plants (39). Notably, the transient burst in expression of genes encoding defense responses at week 26 coincided with conditions that support novel fine root production and ECM colonization, such as the first substantial rainfall of the year and a period of rapid soil warming (SI Appendix, Fig. S1B). In contrast to the elevated expression of stress-associated functions, a significant decrease in transcript abundance was observed in NE for a suite of sugar efflux transporters, primarily the Sugars Will Eventually be Exported Transporters (SWEET) family of transporters (P < 0.001; two-tailed Wilcoxon signed-rank test) (SI Appendix, Fig. S6B). These uniporters regulate the passage of sucrose, glucose, and fructose across cell membranes and are one of the primary exporters of C into the rhizosphere (18, 40). Furthermore, their promoters are targets for microbial transcription activator-like effectors, leading to the appropriation of plant sugar transport during infection (18, 41, 42). The lowered expression of sugar transporters coupled with enhanced defense and effector-triggered immunity responses observed under NE conditions (Fig. 3 and SI Appendix, Fig. S6B) (network modules 2 and 12) represent converging mechanisms by which host trees redefine their economic relationship with the fungal community in response to increased nutrient availability.
Changes in Symbiotic Coordination.
As the extensive seasonal dynamics observed in the tree did not appear to drive community-level fungal processes (Fig. 1B), we carried out a targeted analysis of the three most dominant fungal species to uncover specific signals of symbiotic coordination masked by global analyses. For both P. olivaceum and Co. glaucopus, all of the fungal network modules that were strongly correlated with Norway spruce network modules in NL conditions lost correlation in NE conditions (17 and 18 module comsbinations, respectively; Spearman’s rank correlation coefficient [Rs] > 0.7 or < −0.7) (Fig. 4A). In both genera, these fungal network modules were enriched in functions observed in hemisphere 1, such as signaling, cell cycle processes, and the metabolism of amino acids and secondary metabolites. The most connected Norway spruce module that was decoupled from Co. glaucopus in NE conditions (Norway spruce network module 10) was significantly enriched in plant solute transport functions: for example, NITRATE TRANSPORTER 2 (NRT2), PHOSPHATE TRANSPORTER 1 (PHT1), AMMONIUM TRANSPORTER (AMT2/3-type), multiple phospholipid and fatty acid transporters, and a range of cation transporters associated with Fe, Cu, Ca, and K transport. Notably, this Norway spruce module became coordinated with two P. olivaceum network modules in NE (P. olivaceum network modules 2.1.3 and 2.1.5), which were enriched in fungal functions linked with glycolysis, nucleobase transport, peptidase activity, and the glyoxylate cycle (SI Appendix, Fig. S7). The contrasting responses observed for these two dominant ECM genera could explain P. olivaceum’s persistence in relative community dominance in NE (Fig. 1F). Alternatively, the observed fitness of P. olivaceum in both nutrient conditions could signify the mapping of several closely related Piloderma spp. to the P. olivaceum reference genome. In this scenario, differences in physiology or exploration strategies among these species could explain the perceived fitness of the single reference species.
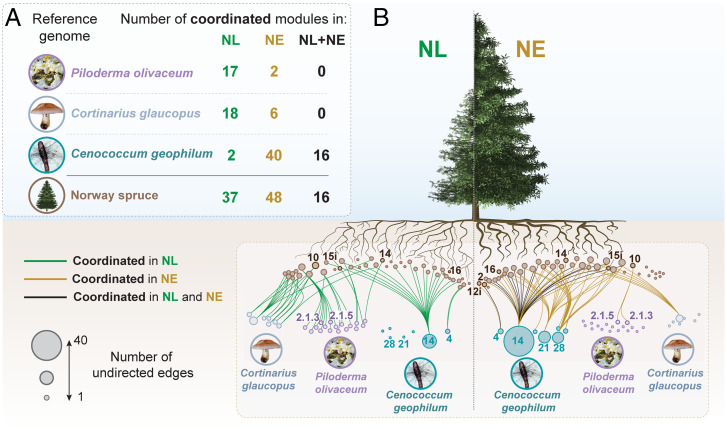
Fig. 4.
Analysis of the coordination between the host-tree and the three most dominant root-associated fungi. (A) Distribution of coordinated network modules (Rs > 0.7 or < −0.7) between Norway spruce and specific fungal species in NL and NE conditions. The number of coordinated network module combinations between Norway spruce and a given fungi are tabulated in the top left of the figure. Those occurring in NL conditions only are shown in green, those occurring in NE conditions only are shown in gold, and those occurring in both treatments are shown in black. (B) Visualization of coordinated Norway spruce and fungal network modules. Edge coloring is consistent with that used in the tabulated section above. The size of each node represents the number of undirected edges to which it is connected. The term “undirected” means the edges represent a two-way relationship between nodes. Network modules that are mentioned in the text have been indicated. Inclusion of an ‘i’ in a network module name indicates the module has the inverse expression profile of the named module. The difference in tree height and aboveground biomass illustrated is modified from Sigurdsson et al. (54) and Majdi and Andersson (117). Functional enrichment analysis for network modules for these species can be found in SI Appendix, Figs. S7 and S8.
Ce. geophilum displayed a fundamentally different response from the two Basidiomycetes, with 89% of the combinations with Norway spruce network modules that were strongly coordinated in NL conditions maintaining this coordination in NE (16 module combinations), while more than double this number became coordinated only in NE (Fig. 4A) (Ce. geophilum). The modules that maintained or increased coordination (primarily Ce. geophilum network modules 4, 14, 21, and 28) were significantly enriched in functions associated with metabolism (fatty acid desaturases, glycogen synthase, and pyrimidine biosynthesis), cell wall remodeling (glucan synthase and endoglucanases), nutrient transport (ammonia and nucleoside permeases), and signal transduction (histidine kinases) (SI Appendix, Fig. S8). Notably, many of the novel module combinations formed in NE conditions included Norway spruce modules that were significantly enriched in plant functions associated with cell wall remodeling, and the biosynthesis and perception of presymbiotic molecular signals: specifically, Norway spruce network modules 14 and 15i, which were coordinated with P. olivaceum in NL but lost coordination in NE conditions, and Norway spruce network module 16. These functions promote reciprocal physical and physiological changes that facilitate symbiosis and included the biosynthesis of terpenes, diterpenes, and strigolactone signal transduction, in addition to a nodulation signaling pathway 1 (NSP1) component, a GRAS transcription factor previously thought to be exclusively linked to nodulation but more recently implicated in mycorrhizal establishment (43, 44).
Coordination between Fungal Effectors and Norway Spruce Genes.
In light of these findings, we targeted the coordination between Norway spruce genes and predicted effectors of the three most dominant fungal species (Fig. 5 and SI Appendix, Supplementary Methods, Figs. S9–S11, and Table S1). Using a combination of predictive tools to establish protein secretion (45, 46) and a machine-learning classifier trained on experimentally supported fungal effectors (47), 33 predicted effectors were identified in P. olivaceum (representing 0.3% of all reads mapped to P. olivaceum), 8 in Co. glaucopus (0.5%), and 23 in Ce. geophilum (0.8%). Putative functions were assigned to the predicted fungal effectors based on the identification of protein domains and transit peptides, which were primarily associated with the modulation of gene expression (transcription factors and subunits of the Mediator complex); redox sensing and signaling; and protein folding, posttranslational modification, and degradation (SI Appendix, Table S1). Intrinsic disorder, a property that causes proteins to lack a fixed three-dimensional structure while retaining biological activity (48), was the most frequently predicted domain identified in the effectors of all three fungal species.
Fig. 5.
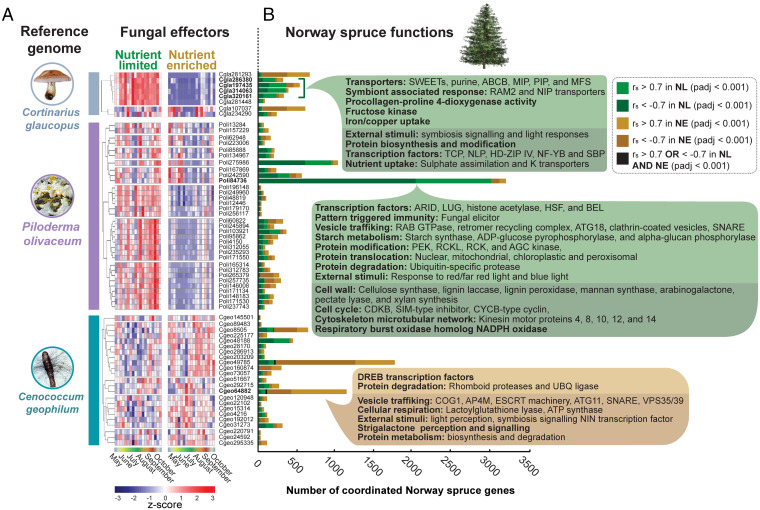
Functional analysis of Norway spruce genes displaying positive and negative coordination with predicted fungal effectors. (A) The present set of genes assigned to Co. glaucopus, P. olivaceum, and Ce. geophilum reference genomes were assessed using the fungal effector predictive tool EffectorP (Materials and Methods). The hierarchically clustered transcript abundance of the predicted fungal effectors is displayed for each species. (B) Spearman’s rank correlation was carried out between the expression profiles of the predicted fungal effectors and all Norway spruce transcripts, in both the NL and NE seasonal time courses. The number of Norway spruce genes that satisfied the given Rs (r > 0.7 or < −0.7) and adjusted P thresholds (<0.001) with a given fungal effector are displayed. Genes positively correlated in NL conditions are displayed in light green, negatively correlated in NL are displayed in dark green, positively correlated in NE are displayed in light gold, and negatively correlated in NE are displayed in dark gold. Significant (adjusted P < 0.05) functional enrichments for these gene groups are provided (SI Appendix, Figs. S9–S11). Specific coordinated effectors discussed in Results are marked in bold.
A number of significantly overrepresented Norway spruce functions were strongly coordinated with the predicted effectors of all three fungal species, regardless of soil nutrient status. These included transcriptional regulation, vesicle trafficking, pattern-triggered immunity (e.g., fungal and bacterial elicitors), and pathogen effector-triggered immunity (e.g., NLR effector receptors, enhanced disease susceptibility 1 [EDS1], and phytoalexin-deficient 4 [PAD4]) (Fig. 5B). In addition, many Norway spruce functions were specifically coordinated with predicted effectors from a single fungal species and showed highly treatment-specific responses. For example, four predicted Co. glaucopus effectors (including putative subunits of the Mediator complex and trafficking protein particle complex) were positively correlated with Norway spruce genes that were significantly enriched in SWEET-family transporters and response-to-symbiont processes (e.g., reduced arbuscular mycorrhization [RAM]2 and Nodulin 26-like intrinsic protein [NIP] transporters) during NL conditions. Notably, for all four predicted effectors, this coordination with sugar efflux and symbiosis responses were lost in NE conditions. A similar loss of coordination in NE soil was observed in a predicted effector of P. olivaceum (Poli84736) containing a cyclin-like subunit (Ssn8) domain of the kinase module of the Mediator complex, which was coordinated with over 3,000 Norway spruce genes in NL conditions—the most of any predicted effector—but only 175 genes in NE soils.
Conversely, the putative effectors of Ce. geophilum demonstrated the greatest level of coordination with Norway spruce genes in NE conditions. For example, a predicted effector (Cgeo64882), containing domains of intrinsic disorder and orthologous to a zinc finger C2H2-type protein in Macrophomina phaseolina, gained 10-times the level of coordination in NE compared to NL conditions. The Norway spruce genes coordinated with this predicted fungal effector were significantly enriched in a large suite of DREB-type transcription factors with homology to specific functionally characterized Arabidopsis thaliana orthologs, which have been linked to the regulation of jasmonic acid biosynthesis (49), repression of disease resistance and mediated cell death (50), and reducing the lignin content of cell walls (51). Taken together, the combination of predictive analytics and high-resolution metatranscriptomics used here provides a strategic approach to identify top candidates for experimental validation, which will dramatically improve our understanding of both the generic and species-specific molecular dialogue occurring between microbial communities and their hosts.
Discussion
Nutrient availability is a critical factor limiting plant productivity and C storage in boreal forest soils. An evolutionary solution to nutrient limitation was the establishment of mutualistic associations, linking plant fitness with root-associated fungal activity. Like most economies, this exchange is sensitive to market forces that can alter the equilibrium of supply and demand (52, 53). Increased nutrient availability, due to anthropogenic N deposition or higher rates of mineralization linked to global warming, is predicted to destabilize this trading status (21), although the severity and lasting consequences of this altered paradigm in boreal symbiosis remains unknown. At the study site used here, 25 y of NE conditions led to a fourfold increase in aboveground tree biomass compared to NL grown trees (54), which coincided with decreases in the flux of photosynthates directed belowground (55), and decreases in autotrophic and heterotrophic soil respiration (56) (SI Appendix, Fig. S1H). We deployed a high-resolution metatranscriptomic analysis to consolidate these environmental observations with the underlying mechanisms impacted by this symbiotic restructuring. We observed functional evidence of host trees redefining the economic status of their relationship with the fungal community via reduction of sugar transporters that are integral to mycorrhizal establishment (57, 58) (SI Appendix, Fig. S6B), coupled with an enhanced defense response involving effector-triggered immunity, downstream signal propagation, and stress-associated transcription factors (Fig. 3, modules 2 and 12). The consequence of these alterations on the fungal community was profound, resulting in a reduced relative abundance of sequencing reads of fungal origin (Fig. 1A), with a notable decrease in the proportion of reads assigned to ECM Basidiomycota in contrast to an increased proportion of melanotic ECM Ascomycota species (Fig. 1C). The sensitivity and resolution of this approach revealed that these changes in community structure were accompanied by, and likely the result of, prominent shifts in broad-scale functional coordination between specific ECM fungi and the host tree (Fig. 4). Perhaps most strikingly, we identified dramatic alterations in the coordination between predicted effectors of the three most prevalent fungal species and over 5,000 Norway spruce transcripts. This finding provides a mechanistic link between the broad structural and functional changes observed in the fungal community, with alterations in the complex molecular dialogue that coordinates the host with its prospective fungal partners.
The response of a single tree to a confluence of effectors from competing fungal species is necessarily an emergent property (59), as individual microbes employ a repertoire of effectors (the “effectome”) that can function pleiotropically (60), redundantly (61), or in concert with each other (62) to affect generic or species-specific processes (63). Global analyses of environmental samples, such as the one described here, can capture these tree-level emergent properties in action, providing crucial insight into their complex in vivo activities and sufficient resolution to identify individual species within the ECM community driving these emergent responses. For Co. glaucopus and P. olivaceum, we observed the transcript abundance of a number of predicted effectors decrease drastically when the resource trading conditions were less favorable in NE conditions, particularly during the period of active fertilization (weeks 23 to 34). In contrast, although the number of Ce. geophilum effectors expressed in NE conditions was the same in NL conditions, the magnitude of their expression was greater in NE conditions (Fig. 5, Ce. geophilum heatmap). Taken together, this suggests the decoupling of P. olivaceum and Co. glaucopus from their host becomes most pronounced following fertilizer application, while the absence of such a response in Ce. geophilum effectors indicates this species is particularly well adapted to N-rich environments.
To date, what little is known of fungal effector function is the result of detailed experimental characterization of individual effectors under controlled conditions. These approaches have provided insights into their diverse modes of action (64), their spatial and temporal kinetics (65), and the host processes commonly targeted (66). Many of the candidate effectors identified in this study possessed qualities linked to interference with protein posttranslational modification (Tyr phosphatase dual domain, protein kinase domain, and Tyr kinase active site), protein stability/folding (DnaJ, calreticulin/calnexin, and peptidyl-prolyl cis-trans isomerase FKBP2/11), or protein degradation (proteasome subunit-β5 and UBQ-conjugating E2). Such mechanisms have been widely described during phytopathogenic interactions (67), for example, during infection the Phytophthora sojae effector Avr1d competitively binds with a soybean E3 ubiquitin ligase, preventing its self-ubiquitination and degradation, leading to increased plant susceptibility to infection (68). Similarly, in Pseudomonas syringae, the effector HopBF1 phosphorylates plant HSP90 to inhibit the chaperone’s ATPase activity, which blocks its initiation of the plant hypersensitive response (69).
We also identified five candidate effectors putatively involved in host transcriptional regulation [e.g., leucine zipper domain, Zn (2)-C6 fungal-type DNA-binding domain, and subunits of the Mediator complex and transcription initiation factor IID]. Compromised immunity arising from effector-mediated interference of host transcription has been described in several plant–microbe interactions, including mutualistic symbioses. For example, the interaction of the symbiosis effectors SP7 of Rhizophagus irregularis with the pathogenesis-related transcription factor ERF19 of Medicago truncatula, and MiSSP7 and MiSSP7.6 of Laccaria bicolor with the jasmonic acid transcriptional corepressor PtJAZ6 and the transcription factors PtTrihelix1 of Populus trichocarpa, respectively (16, 70, 71). Notably, we identified a number of effectors with potential targeting to mitochondria, two of which (one from Co. glaucopus and one from P. olivaceum) possess a domain common to translocase complex proteins of the inner membrane (TIM), which has been linked to mitochondrial-mediated plant immunity in A. thaliana (72, 73). Although mitochondria play a central role in the plant defense response, it is only recently that the effector proteins MoCDIP4 and Avr-Pita from Magnaporthe oryzae were shown to modulate rice susceptibility by targeting a mitochondrial DnaJ protein (74) and a cytochrome c oxidase (75), respectively, revealing a novel pathogen infection strategy targeting host mitochondria. Future efforts to experimentally validate candidate fungal effectors (such as those identified here) (Fig. 5 and SI Appendix, Table S1) would be bolstered by the integration of these functional predictions with the foreknowledge of their expression dynamics, not only in regard to treatment and colonization-checkpoints, but in relation to the greater fungal effectome (including self and fungal competitors) and their coexpression with host functions.
The reduction of total fungal reads in NE soils (consistent across growing seasons and methodologies) (Fig. 1A and SI Appendix, Fig. S2B) concurs with previous observations that increasing nutrient gradients or active fertilization in boreal soils decreases ECM mycelium and sporocarp production (76, 77). Extramatrical mycelia represent a substantial proportion of total belowground C (78). However, beyond absolute fungal biomass, mycelial taxonomic composition has also been shown to influence the accumulation of C through the production of degradative enzymes (79) and the varying recalcitrance of fungal necromass (25, 80, 81). The Basidiomycota Cortinarius underwent the most dramatic reduction in assigned fungal reads in response to NE conditions (Fig. 1 and SI Appendix, Fig. S2), coincident with a decoupling of broad-scale fungal and tree functions related to growth and solute transport (Fig. 4), as well as highly specific predicted fungal effectors and tree functions associated with supporting mycorrhizal partners (e.g., sugar efflux transporters and symbiont-response processes) (Fig. 5). This genus comprises well characterized ECM fungi with energetically demanding lifestyles that employ medium-distance fringe exploration strategies and secrete oxidative enzymes capable of degrading lignin (79, 82–85). Due to the elaborate foraging strategies of Cortinarius spp., they have some of the highest C demands of all ECM (86) and are thus highly sensitive to reduced C flow from host roots (83, 87–89). The capacity of members of the genus Cortinarius to degrade recalcitrant soil organic matter is unique among ECM fungi and they exert a disproportionate impact on soil C turnover (90). A recent study demonstrated the presence of Cortinarius species-complex members in the organic topsoil of boreal forests resulted in a 33% decrease in local C storage (90). This finding disputes the prevailing consensus that the functional redundancy inherent to complex microbial communities will offset the loss of an individual species. Thus, the collapse in symbiotic coordination and diminished presence of Cortinarius spp. in the fungal community observed in our study suggests that increased atmospheric N deposition and enhanced mineralization rates linked to warming soils will reduce the abundance and activity of these keystone degraders of lignified plant matter and humus in boreal soils.
Contrasting this reduction in Basidiomycete abundance, NE resulted in increases in the Ascomycetes Ce. geophilum (Fig. 1), accompanied by an intensification in transcriptomic coordination with the host tree, at both a broad-scale with fungal processes linked to metabolism, cell wall remodeling, and nutrient transport (Fig. 4 and SI Appendix, Fig. S8), and a fine-scale involving a suite of predicted fungal effectors with putative functions, ranging from transcriptional regulation to protein ubiquitylation (Fig. 5 and SI Appendix, Table S1). Ce. geophilum is a globally distributed and highly versatile short-range exploratory ECM fungus with distinctive black hyphae resulting from heavily melanized cell walls (91, 92). This heavy melanization makes these fungi resilient to adverse environmental conditions, such as drought (93, 94), and can act as virulence factors during root colonization (95, 96). Notably, melanized necromass is also highly recalcitrant to decomposition (97, 98), with the mycelia of Ce. geophilum demonstrated to persist in soils up to 10 times longer than that of other ECM species (92, 99). This suggests that while NE resulted in reduced overall fungal biomass (31), it triggered an absolute increase in recalcitrant fungal necromass that is more likely to contribute to long-term C storage. This store of organic C would necessarily benefit microorganisms with the capacity to access it. Functional analysis of Ce. geophilum transcripts highly expressed in both nutrient treatments revealed a significant enrichment of chitinases and functions associated with polycyclic aromatic hydrocarbon degradation (SI Appendix, Fig. S4) (Ce. geophilum), a cohort of functions that have previously been implicated in the degradation of environmental pollutants and, notably, melanins (100). These enzymes play essential roles in accessing stores of melanized fungal necromass and the self-digestion processes (autolysis) utilized for remodeling of mycelial architecture (80). The expression of these functions in both NL and NE conditions suggests that the capability to oxidatively degrade melanin could be a crucial function for Ce. geophilum, regardless of soil nutrient status, with the corollary that access to these growing reserves of recalcitrant C in NE soil has the potential to further reinforce this species’ position in the rhizospheric community. However, strategic experimentation will be needed to discriminate the overlapping roles these degradative enzymes play in mycelial autolytic remodeling and nutrient scavenging.
As boreal forests harbor globally significant amounts of C, understanding the biogeochemistry of boreal soils is essential for informing strategic responses to future climate scenarios. One area where such understanding is needed was revealed by a recent metaanalysis of over 100 elevated CO2 experiments (12) that has challenged the consensus of existing ecosystem modeling. Previous models had predicted increases in terrestrial C sequestration under elevated CO2 conditions, while this metaanalysis suggests instead that the increased biomass of ECM-associated plants will be supported by augmented nutrient uptake and result in reduced soil organic C accumulation. Recent field experiments have shown that trees exposed to elevated CO2 increased belowground C allocation, resulting in enhanced production of fine roots (101) and higher rates of soil CO2 efflux (102). This alteration in belowground C allocation represents a revealing counterpoint to the impact of soil nutrient enrichment examined in the present study.
CO2-driven increases in belowground C-availability could cause reciprocal changes in the dominance and activity of Basidiomycetes (such as Cortinarius and Piloderma species) at the expense of Ascomycetes (such as Ce. geophilum and Meliniomyces species). Potentially, this CO2-driven shift in community composition could result in a gradual increase in oxidative decomposition of soil organic matter (103) and a decrease in recalcitrant fungal necromass, triggering a reduction in the capacity of boreal forest soils to sequester C but an increase in tree biomass. This conflict between observed and modeled C cycling highlights the uncertainty of climate projections that underestimate the importance and scale of rhizospheric processes and emphasizes the need to monitor and integrate the diverse biogeochemical processes contributed by microbial communities and their host plants. Metatranscriptomic approaches, such as the one described here, can aid in functionally resolving these processes and will add to our conceptual framework of global C sequestration and ecosystem stability in future climate conditions. Notably, this approach advances studies of complex microbial communities from being purely taxonomic, using DNA-based methods, to identifying causative process-driven links and alterations in the transcriptional coordination of mycorrhizal associations. Additionally, minor user adjustments to this opensource and reproducible methodology will enable high-resolution monitoring of a range of host–microbe systems, from pathologies arising from dysbiosis in the human body, to applications in a diversity of threatened ecosystems.
Materials and Methods
Samples were collected at the Flakaliden research site (64°07′N, 19°27′E, altitude 310 to 320 m) in the boreal forests of northern Sweden [for details of the site, please refer to Haas et al. (83)]. Treatment of designated experimental plots within the forest stand of an optimal nutrient solution of macro- and micronutrients—including N, P, K, Mg, Ca, S, Fe, B, Mn, Cu, and Zn—commenced in 1987 and was updated yearly based on analysis of the foliar nutrient status at the site (104). Five root sample replicates were harvested from a single NL plot and a single NE plot (SI Appendix, Fig. S1A, plots 6A and 7A, respectively) at each time point during 19 wk over the 2011 growing season (May to October). Root samples were harvested from 0- to 25-cm-deep soil, cleaned, frozen, and ground. RNA was isolated from 75 mg of tissue using the Ambion Plant RNA Isolation Aid and the Ambion RNAqueous kit, followed by a DNA-free kit DNase treatment. RNA concentration was determined using a Nanodrop NL100 and RNA quality was assessed using an Agilent 2100 Bioanalyzer. The RNA-seq raw data are publicly available in the European nucleotide archive (ENA) with the accession no. PRJEB35805 (105). In 2012, three sample replicates were harvested from each of three replicate NL plots and three replicate NE plots (SI Appendix, Fig. S1A, plots 4A, 6A, and 14A; and 1A, 7A, 13A, respectively), at four time points over the 2012 growing season. For these samples, DNA and RNA was isolated. The ITS1 amplicon sequencing data generated from these samples were published previously (83) and reanalyzed by Schneider et al. (106). RNA extracted from these Norway spruce root samples was sequenced as described above. This low temporal resolution collection was used to validate the high temporal resolution samples collected in 2011, in reference to 1) year-to-year variation, 2) interplot variation, and 3) a complimentary ITS amplicon sequencing approach.
Norway spruce RNA-seq data were preprocessed and aligned with SortmeRNA (107), Trimmomatic (108), and Salmon (109). Preprocessing and analysis of metatranscriptomic data were implemented in a reproducible snakemake workflow available on Bitbucket (https://bitbucket.org/scilifelab-lts/n_street_1801/). A detailed description of software, including parameters and versions used in the workflow, has been published in a separate article (106). Assembled fungal transcripts were assigned taxonomic annotation based on sequence similarity to available reference genomes, with ≥85% similarity required for species assignment, ≥60% similarity for genus assignment, and ≥45% for phylum assignment (110, 111). If a similarity threshold to a given taxonomic level was not satisfied, the prefix “Unclassified” was assigned before the next highest classification (e.g., “Unclassified Cortinarius” indicates similarity to the available reference genome was >60% but <85%). Functional annotations were assigned using eggnog-mapper (112) and the eggNOG database. For both Norway spruce and fungi, high-dimensional data analysis was performed using principal component analysis (PCA) and extended with PHATE (113). Differential expression analysis and variance stabilization transformation were carried out with DESeq2 (114). Coexpression networks were constructed using Seidr (115) and clustered using Infomap (116). Gene-enrichment tests were carried out with Gofer3, an in-house tool at the Umeå Plant Science Centre (DOI:10.5281/zenodo.3731544). All enrichment tests can be accessed on the BRA web app (https://www.boreal-atlas.info/). All other analyses were executed using in-house tools. All scripts are available on Github (https://github.com/loalon/flakaliden-2011).
Supplementary Material
Acknowledgments
We thank Mathieu Castelain for his technical assistance during field sampling and subsequent RNA extraction; the Umeå Plant Science Centre bioinformatics facility (https://www.upsc.se/platforms/upsc-bioinformatics-facility.html) for technical support with regards to the RNA-sequencing data preprocessing and analyses; and Anna Rosling and Åsa Strand for their comments on the manuscript. T.N., N.R.S., and V.H. acknowledge funding from the Swedish University of Agricultural Science’s Trees and Crops for the Future (TC4F) program. We acknowledge support from the National Genomics Infrastructure in Stockholm funded by Science for Life Laboratory, the Knut and Alice Wallenberg Foundation and the Swedish Research Council, and Swedish National Infrastructure for Computing/Uppsala Multidisciplinary Center for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2118852119/-/DCSupplemental.
Data Availability
All enrichment tests can be accessed on the BRA web app (https://www.boreal-atlas.info/). The RNA-seq raw data are publicly available in the European nucleotide archive (ENA), https://www.ebi.ac.uk/ena/browser/ (accession no. PRJEB35805) (105). All other analyses were executed using in-house tools. All Scripts data have been deposited in GitHub. (https://github.com/loalon/flakaliden-2011).
References
- 1.Trivedi P., Leach J. E., Tringe S. G., Sa T., Singh B. K., Plant-microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 18, 607–621 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Eckburg P. B., et al. , Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venter J. C., et al. , Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Tringe S. G., et al. , Comparative metagenomics of microbial communities. Science 308, 554–557 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Belstrøm D., et al. , Metagenomic and metatranscriptomic analysis of saliva reveals disease-associated microbiota in patients with periodontitis and dental caries. NPJ Biofilms Microbiomes 3, 23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lloyd-Price J., et al. ; IBDMDB Investigators, Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amato P., et al. , Metatranscriptomic exploration of microbial functioning in clouds. Sci. Rep. 9, 4383 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crits-Christoph A., Diamond S., Butterfield C. N., Thomas B. C., Banfield J. F., Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis. Nature 558, 440–444 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Pérez-Losada M., Castro-Nallar E., Bendall M. L., Freishtat R. J., Crandall K. A., Dual transcriptomic profiling of host and microbiota during health and disease in pediatric asthma. PLoS One 10, e0131819 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Starr E. P., Nuccio E. E., Pett-Ridge J., Banfield J. F., Firestone M. K., Metatranscriptomic reconstruction reveals RNA viruses with the potential to shape carbon cycling in soil. Proc. Natl. Acad. Sci. U.S.A. 116, 25900–25908 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tedersoo L., Bahram M., Mycorrhizal types differ in ecophysiology and alter plant nutrition and soil processes. Biol. Rev. Camb. Philos. Soc. 94, 1857–1880 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Terrer C., et al. , A trade-off between plant and soil carbon storage under elevated CO2. Nature 591, 599–603 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Brundrett M. C., Mycorrhizal associations and other means of nutrition of vascular plants: Understanding the global diversity of host plants by resolving conflicting information and developing reliable means of diagnosis. Plant Soil 320, 37–77 (2009). [Google Scholar]
- 14.van der Heijden M. G. A., Martin F. M., Selosse M. A., Sanders I. R., Mycorrhizal ecology and evolution: The past, the present, and the future. New Phytol. 205, 1406–1423 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Kohler A., et al. ; Mycorrhizal Genomics Initiative Consortium, Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat. Genet. 47, 410–415 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Plett J. M., et al. , Effector MiSSP7 of the mutualistic fungus Laccaria bicolor stabilizes the Populus JAZ6 protein and represses jasmonic acid (JA) responsive genes. Proc. Natl. Acad. Sci. U.S.A. 111, 8299–8304 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L. H., et al. , A diverse member of the fungal Avr4 effector family interacts with de-esterified pectin in plant cell walls to disrupt their integrity. Sci. Adv. 7, eabe0809 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L. Q., et al. , Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–532 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith S. E., Read D. J., Mycorrhizal Symbiosis (Academic Press, London, UK, ed. 3, 2008). [Google Scholar]
- 20.Hogberg P., Nasholm T., Franklin O., Hogberg M. N., Tamm review: On the nature of the nitrogen limitation to plant growth in Fennoscandian boreal forests. For. Ecol. Manage. 403, 161–185 (2017). [Google Scholar]
- 21.Kicklighter D. W., Melillo J. M., Monier E., Sokolov A. P., Zhuang Q., Future nitrogen availability and its effect on carbon sequestration in Northern Eurasia. Nat. Commun. 10, 3024 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maaroufi N. I., et al. , Anthropogenic nitrogen deposition enhances carbon sequestration in boreal soils. Glob. Change Biol. 21, 3169–3180 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Hogberg P., Fan H. B., Quist M., Binkley D., Tamm C. O., Tree growth and soil acidification in response to 30 years of experimental nitrogen loading on boreal forest. Glob. Change Biol. 12, 489–499 (2006). [Google Scholar]
- 24.Gundale M. J., From F., Bach L. H., Nordin A., Anthropogenic nitrogen deposition in boreal forests has a minor impact on the global carbon cycle. Glob. Change Biol. 20, 276–286 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Clemmensen K. E., et al. , Roots and associated fungi drive long-term carbon sequestration in boreal forest. Science 339, 1615–1618 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Ruckstuhl K. E., Johnson E. A., Miyanishi K., Introduction. The boreal forest and global change. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 2245–2249 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soja A. J., et al. , Climate-induced boreal forest change: Predictions versus current observations. Glob. Planet. Change 56, 274–296 (2007). [Google Scholar]
- 28.Averill C., Turner B. L., Finzi A. C., Mycorrhiza-mediated competition between plants and decomposers drives soil carbon storage. Nature 505, 543–545 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Craig H., et al. , Nitrogen addition alters composition, diversity, and functioning of microbial communities in mangrove soils: An incubation experiment. Soil Biol. Biochem. 153, 108076 (2021). [Google Scholar]
- 30.Leff J. W., et al. , Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc. Natl. Acad. Sci. U.S.A. 112, 10967–10972 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maaroufi N. I., et al. , Nutrient optimization of tree growth alters structure and function of boreal soil food webs. For. Ecol. Manage. 428, 46–56 (2018). [Google Scholar]
- 32.Shu B., Li W., Liu L., Wei Y., Shi S., Transcriptomes of arbuscular mycorrhizal fungi and litchi host interaction after tree girdling. Front. Microbiol. 7, 408 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dyrka W., et al. , Diversity and variability of NOD-like receptors in fungi. Genome Biol. Evol. 6, 3137–3158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kataoka R., Taniguchi T., Ooshima H., Futai K., Comparison of the bacterial communities established on the mycorrhizae formed on Pinus thunbergii root tips by eight species of fungi. Plant Soil 304, 267–275 (2008). [Google Scholar]
- 35.Krywolap G. N., Casida L. E. Jr., An antibiotic produced by the mycorrhizal fungus Cenococcum graniforme. Can. J. Microbiol. 10, 365–370 (1964). [DOI] [PubMed] [Google Scholar]
- 36.Ashburner M., et al. ; The Gene Ontology Consortium, Gene ontology: Tool for the unification of biology. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grossmann S., Bauer S., Robinson P. N., Vingron M., Improved detection of overrepresentation of gene-ontology annotations with parent child analysis. Bioinformatics 23, 3024–3031 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Usadel B., et al. , A guide to using MapMan to visualize and compare Omics data in plants: A case study in the crop species, Maize. Plant Cell Environ. 32, 1211–1229 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Gutha L. R., Reddy A. R., Rice DREB1B promoter shows distinct stress-specific responses, and the overexpression of cDNA in tobacco confers improved abiotic and biotic stress tolerance. Plant Mol. Biol. 68, 533–555 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Hennion N., et al. , Sugars en route to the roots. Transport, metabolism and storage within plant roots and towards microorganisms of the rhizosphere. Physiol. Plant. 165, 44–57 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Chu Z., et al. , Targeting xa13, a recessive gene for bacterial blight resistance in rice. Theor. Appl. Genet. 112, 455–461 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Yang B., Sugio A., White F. F., Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. U.S.A. 103, 10503–10508 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delaux P. M., Bécard G., Combier J. P., NSP1 is a component of the Myc signaling pathway. New Phytol. 199, 59–65 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Bonfante P., Genre A., Mechanisms underlying beneficial plant-fungus interactions in mycorrhizal symbiosis. Nat. Commun. 1, 48 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Almagro Armenteros J. J., et al. , SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Käll L., Krogh A., Sonnhammer E. L., Advantages of combined transmembrane topology and signal peptide prediction—The Phobius web server. Nucleic Acids Res. 35, W429–W432 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sperschneider J., Dodds P. N., Gardiner D. M., Singh K. B., Taylor J. M., Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol. Plant Pathol. 19, 2094–2110 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang L. N., et al. , The Phytophthora infestans AVR2 effector escapes R2 recognition through effector disordering. Mol. Plant Microbe Interact. 33, 921–931 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Chen H. Y., et al. , ORA47 (octadecanoid-responsive AP2/ERF-domain transcription factor 47) regulates jasmonic acid and abscisic acid biosynthesis and signaling through binding to a novel cis-element. New Phytol. 211, 599–613 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Zhao Z. X., et al. , RPW8.1 enhances the ethylene-signaling pathway to feedback-attenuate its mediated cell death and disease resistance in Arabidopsis. New Phytol. 229, 516–531 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakata M. T., Sakamoto S., Nuoendagula S., Kajita S., Mitsuda N., Fiber cell-specific expression of the VP16-fused ethylene response factor 41 protein increases biomass yield and alters lignin composition. Front. Plant Sci. 12, 654655 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Franklin O., Näsholm T., Högberg P., Högberg M. N., Forests trapped in nitrogen limitation—An ecological market perspective on ectomycorrhizal symbiosis. New Phytol. 203, 657–666 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Högberg M. N., Högberg P., Wallander H., Nilsson L. O., Carbon-nitrogen relations of ectomycorrhizal mycelium across a natural nitrogen supply gradient in boreal forest. New Phytol. 232, 1839–1848 (2021). [DOI] [PubMed] [Google Scholar]
- 54.Sigurdsson B. D., Medhurst J. L., Wallin G., Eggertsson O., Linder S., Growth of mature boreal Norway spruce was not affected by elevated [CO(2)] and/or air temperature unless nutrient availability was improved. Tree Physiol. 33, 1192–1205 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Iivonen S., Kaakinen S., Jolkkonen A., Vapaavuori E., Linder S., Influence of long-term nutrient optimization on biomass, carbon, and nitrogen acquisition and allocation in Norway spruce. Can. J. For. Res. 36, 1563–1571 (2006). [Google Scholar]
- 56.Olsson P., Linder S., Giesler R., Hogberg P., Fertilization of boreal forest reduces both autotrophic and heterotrophic soil respiration. Glob. Change Biol. 11, 1745–1753 (2005). [Google Scholar]
- 57.Manck-Götzenberger J., Requena N., Arbuscular mycorrhiza symbiosis induces a major transcriptional reprogramming of the potato SWEET sugar transporter family. Front. Plant Sci. 7, 487 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao S., et al. , Transcriptomic analysis reveals the possible roles of sugar metabolism and export for positive mycorrhizal growth responses in soybean. Physiol. Plant. 166, 712–728 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Arroyo-Velez N., González-Fuente M., Peeters N., Lauber E., Noël L. D., From effectors to effectomes: Are functional studies of individual effectors enough to decipher plant pathogen infectious strategies? PLoS Pathog. 16, e1009059 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu H., Rao X., Zhang K., Nucleoside diphosphate kinase (Ndk): A pleiotropic effector manipulating bacterial virulence and adaptive responses. Microbiol. Res. 205, 125–134 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Kvitko B. H., et al. , Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 5, e1000388 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei H. L., Zhang W., Collmer A., Modular study of the type III effector repertoire in Pseudomonas syringae pv. tomato DC3000 Reveals a matrix of effector interplay in pathogenesis. Cell Rep. 23, 1630–1638 (2018). [DOI] [PubMed] [Google Scholar]
- 63.de Freitas Pereira M., et al. , Secretome analysis from the ectomycorrhizal ascomycete Cenococcum geophilum. Front. Microbiol. 9, 141 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Plett J. M., et al. , Mycorrhizal effector PaMiSSP10b alters polyamine biosynthesis in Eucalyptus root cells and promotes root colonization. New Phytol. 228, 712–727 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Depotter J. R. L., et al. , Effectors with different gears: Divergence of Ustilago maydis effector genes is associated with their temporal expression pattern during plant infection. J. Fungi (Basel) 7, 16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Daguerre Y., et al. , The mutualism effector MiSSP7 of Laccaria bicolor alters the interactions between the poplar JAZ6 protein and its associated proteins. Sci. Rep. 10, 20362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.He Q., McLellan H., Boevink P. C., Birch P. R. J., All roads lead to susceptibility: The many modes of action of fungal and oomycete intracellular effectors. Plant Commun. 1, 100050 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin Y., et al. , Phytophthora sojae effector Avr1d functions as an E2 competitor and inhibits ubiquitination activity of GmPUB13 to facilitate infection. Proc. Natl. Acad. Sci. U.S.A. 118, e2018312118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lopez V. A., et al. , A bacterial effector mimics a host HSP90 client to undermine immunity. Cell 179, 205–218.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kloppholz S., Kuhn H., Requena N., A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr. Biol. 21, 1204–1209 (2011). [DOI] [PubMed] [Google Scholar]
- 71.Kang H., et al. , The small secreted effector protein MiSSP7.6 of Laccaria bicolor is required for the establishment of ectomycorrhizal symbiosis. Environ. Microbiol. 22, 1435–1446 (2020). [DOI] [PubMed] [Google Scholar]
- 72.Lister R., et al. , A transcriptomic and proteomic characterization of the Arabidopsis mitochondrial protein import apparatus and its response to mitochondrial dysfunction. Plant Physiol. 134, 777–789 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang Y., et al. , Mitochondrial AtPAM16 is required for plant survival and the negative regulation of plant immunity. Nat. Commun. 4, 2558 (2013). [DOI] [PubMed] [Google Scholar]
- 74.Xu G., et al. , A fungal effector targets a heat shock-dynamin protein complex to modulate mitochondrial dynamics and reduce plant immunity. Sci. Adv. 6, eabb7719 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Han J., et al. , The fungal effector Avr-pita suppresses innate immunity by increasing COX activity in rice mitochondria. Rice (N. Y.) 14, 12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nilsson L. O., Wallander H., Production of external mycelium by ectomycorrhizal fungi in a Norway spruce forest was reduced in response to nitrogen fertilization. New Phytol. 158, 409–416 (2003). [Google Scholar]
- 77.Bahr A., et al. , Growth of ectomycorrhizal fungal mycelium along a Norway spruce forest nitrogen deposition gradient and its effect on nitrogen leakage. Soil Biol. Biochem. 59, 38–48 (2013). [Google Scholar]
- 78.Wallander H., et al. , Evaluation of methods to estimate production, biomass and turnover of ectomycorrhizal mycelium in forests soils—A review. Soil Biol. Biochem. 57, 1034–1047 (2013). [Google Scholar]
- 79.Bödeker I. T., Nygren C. M., Taylor A. F., Olson A., Lindahl B. D., ClassII peroxidase-encoding genes are present in a phylogenetically wide range of ectomycorrhizal fungi. ISME J. 3, 1387–1395 (2009). [DOI] [PubMed] [Google Scholar]
- 80.Ekblad A., et al. , The production and turnover of extramatrical mycelium of ectomycorrhizal fungi in forest soils: Role in carbon cycling. Plant Soil 366, 1–27 (2013). [Google Scholar]
- 81.Wilkinson A., Alexander I. J., Johnson D., Species richness of ectomycorrhizal hyphal necromass increases soil CO2 efflux under laboratory conditions. Soil Biol. Biochem. 43, 1350–1355 (2011). [Google Scholar]
- 82.Allison S. D., Czimczik C. I., Treseder K. K., Microbial activity and soil respiration under nitrogen addition in Alaskan boreal forest. Glob. Change Biol. 14, 1156–1168 (2008). [Google Scholar]
- 83.Haas J. C., et al. , Microbial community response to growing season and plant nutrient optimisation in a boreal Norway spruce forest. Soil Biol. Biochem. 125, 197–209 (2018). [Google Scholar]
- 84.Lilleskov E. A., Hobbie E. A., Horton T. R., Conservation of ectomycorrhizal fungi: Exploring the linkages between functional and taxonomic responses to anthropogenic N deposition. Fungal Ecol. 4, 174–183 (2011). [Google Scholar]
- 85.Heinonsalo J., et al. , Evidences on the ability of mycorrhizal genus Piloderma to use organic nitrogen and deliver it to scots pine. PLoS One 10, e0131561 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hobbie E. A., Carbon allocation to ectomycorrhizal fungi correlates with belowground allocation in culture studies. Ecology 87, 563–569 (2006). [DOI] [PubMed] [Google Scholar]
- 87.Yarwood S. A., Myrold D. D., Högberg M. N., Termination of belowground C allocation by trees alters soil fungal and bacterial communities in a boreal forest. FEMS Microbiol. Ecol. 70, 151–162 (2009). [DOI] [PubMed] [Google Scholar]
- 88.Clemmensen K. E., et al. , Carbon sequestration is related to mycorrhizal fungal community shifts during long-term succession in boreal forests. New Phytol. 205, 1525–1536 (2015). [DOI] [PubMed] [Google Scholar]
- 89.Bonner M. T. L., et al. , Why does nitrogen addition to forest soils inhibit decomposition? Soil Biol. Biochem. 137, 107570 (2019). [Google Scholar]
- 90.Lindahl B. D., et al. , A group of ectomycorrhizal fungi restricts organic matter accumulation in boreal forest. Ecol. Lett. 24, 1341–1351 (2021). [DOI] [PubMed] [Google Scholar]
- 91.Peter M., et al. , Ectomycorrhizal ecology is imprinted in the genome of the dominant symbiotic fungus Cenococcum geophilum. Nat. Commun. 7, 12662 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fernandez C. W., McCormack M. L., Hill J. M., Pritchard S. G., Koide R. T., On the persistence of Cenococcum geophilum ectomycorrhizas and its implications for forest carbon and nutrient cycles. Soil Biol. Biochem. 65, 141–143 (2013). [Google Scholar]
- 93.Fernandez C. W., Koide R. T., The function of melanin in the ectomycorrhizal fungus Cenococcum geophilum under water stress. Fungal Ecol. 6, 479–486 (2013). [Google Scholar]
- 94.Gadd G. M., Derome L., Biosorption of copper by fungal melanin. Appl. Microbiol. Biotechnol. 29, 610–617 (1988). [Google Scholar]
- 95.Bell A. A., Wheeler M. H., Biosynthesis and functions of fungal melanins. Annu. Rev. Phytopathol. 24, 411–451 (1986). [Google Scholar]
- 96.Cordero R. J. B., Casadevall A., Functions of fungal melanin beyond virulence. Fungal Biol. Rev. 31, 99–112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fernandez C. W., Koide R. T., Initial melanin and nitrogen concentrations control the decomposition of ectomycorrhizal fungal litter. Soil Biol. Biochem. 77, 150–157 (2014). [Google Scholar]
- 98.Fernandez C. W., Kennedy P. G., Melanization of mycorrhizal fungal necromass structures microbial decomposer communities. J. Ecol. 106, 468–479 (2018). [Google Scholar]
- 99.Gray L., Kernaghan G., Fungal succession during the decomposition of ectomycorrhizal fine roots. Microb. Ecol. 79, 271–284 (2020). [DOI] [PubMed] [Google Scholar]
- 100.Borovanský J., Elleder M., Melanosome degradation: Fact or fiction. Pigment Cell Res. 16, 280–286 (2003). [DOI] [PubMed] [Google Scholar]
- 101.Jackson R. B., Cook C. W., Pippen J. S., Palmer S. M., Increased belowground biomass and soil CO2 fluxes after a decade of carbon dioxide enrichment in a warm-temperate forest. Ecology 90, 3352–3366 (2009). [DOI] [PubMed] [Google Scholar]
- 102.Bernhardt E. S., et al. , Long-term effects of free air CO2 enrichment (FACE) on soil respiration. Biogeochemistry 77, 91–116 (2006). [Google Scholar]
- 103.Adamczyk B., et al. , Plant roots increase both decomposition and stable organic matter formation in boreal forest soil. Nat. Commun. 10, 3982 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Linder S., Foliar analysis for detecting and correcting nutrient imbalances in Norway spruce. Ecol. Bull. 44, 178–190 (1995). [Google Scholar]
- 105.Law S. R., et al. , Data from “Metatranscriptomics captures dynamic shifts in mycorrhizal coordination in boreal forests.” European Nucleotide Archive. https://www.ebi.ac.uk/ena/browser/text-search?query=PRJEB35805. Deposited 1 December 2020. [DOI] [PMC free article] [PubMed]
- 106.Schneider A. N., et al. , Comparative fungal community analyses using metatranscriptomics and internal transcribed spacer amplicon sequencing from Norway spruce. mSystems 6, e00884-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kopylova E., Noé L., Touzet H., SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–3217 (2012). [DOI] [PubMed] [Google Scholar]
- 108.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Luo C., Rodriguez-R L. M., Konstantinidis K. T., MyTaxa: An advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Res. 42, e73 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Alneberg J., et al. , BARM and BalticMicrobeDB, a reference metagenome and interface to meta-omic data for the Baltic Sea. Sci. Data 5, 180146 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cantalapiedra C. P., Hernández-Plaza A., Letunic I., Bork P., Huerta-Cepas J., eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38, 5825–5829 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Moon K. R., et al. , Visualizing structure and transitions in high-dimensional biological data. Nat. Biotechnol. 37, 1482–1492 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schiffthaler B., van Zalen E., Serrano A. R., Street N. R., Delhomme N., Seidr: A gene meta-network calculation toolkit. bioRxiv [Preprint] (2018). http 10.1101/250696 (Accessed 1 March 2018). [DOI]
- 116.Rosvall M., Axelsson D., Bergstrom C. T., The map equation. Eur. Phys. J. Spec. Top. 178, 13–23 (2009). [Google Scholar]
- 117.Majdi H., Andersson P., Fine root production and turnover in a Norway spruce stand in northern Sweden: Effects of nitrogen and water manipulation. Ecosystems 8, 191–199 (2005). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All enrichment tests can be accessed on the BRA web app (https://www.boreal-atlas.info/). The RNA-seq raw data are publicly available in the European nucleotide archive (ENA), https://www.ebi.ac.uk/ena/browser/ (accession no. PRJEB35805) (105). All other analyses were executed using in-house tools. All Scripts data have been deposited in GitHub. (https://github.com/loalon/flakaliden-2011).