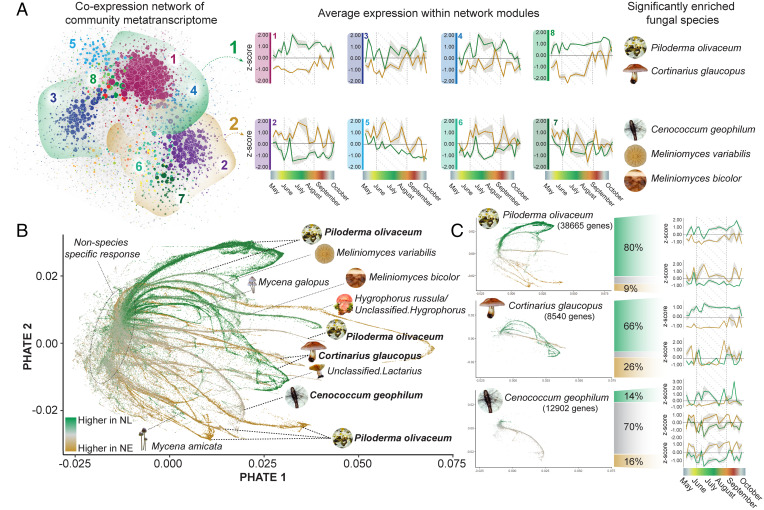
Fig. 2.
Remodeling of fungal community metatranscriptome in samples from NL and NE plots from the same forest site over a growing season. (A) Visualization of the root-associated fungal community network of samples collected from NL and NE forest plots over a growing season. Average expression profiles of the eight largest network modules are displayed, with NL in green and NE in gold, and the SD indicated by gray shading. Significantly enriched functional categories (adjusted P < 0.05; Fisher exact test) for each network module can be found in SI Appendix, Fig. S3. The network modules were grouped into two hemispheres, with hemisphere 1 (indicated in green) displaying higher average expression in NL conditions, and hemisphere 2 (indicated in gold) displaying higher average expression in NE conditions. Fungal species that are significantly enriched in each hemisphere (P < 0.0001; one-proportion z-test) are indicated. (B) Low-dimensional visualization of all fungal genes in both NL and NE conditions. The gene nodes have been colored based on the fold-change differences between NL and NE conditions. Where possible, the branching arms have been assigned to distinct taxonomic assignments, with species names reflecting the reference genome to which RNA-seq reads were assigned. (C) The genes of the three fungal species with the highest assignment of RNA-seq reads (mapped to a specific reference genome) were isolated and separated based on three criteria: 1) fold-change was <−0.5 (higher abundance in NL), indicated in green; 2) fold-change was >0.5 (higher abundance in NE), indicated in gold; and 3) the fold-change was < 0.5 but >−0.5. The average expression profiles of the genes meeting these three criteria have been visualized and corresponding functional enrichment analysis can be found in SI Appendix, Fig. S4.