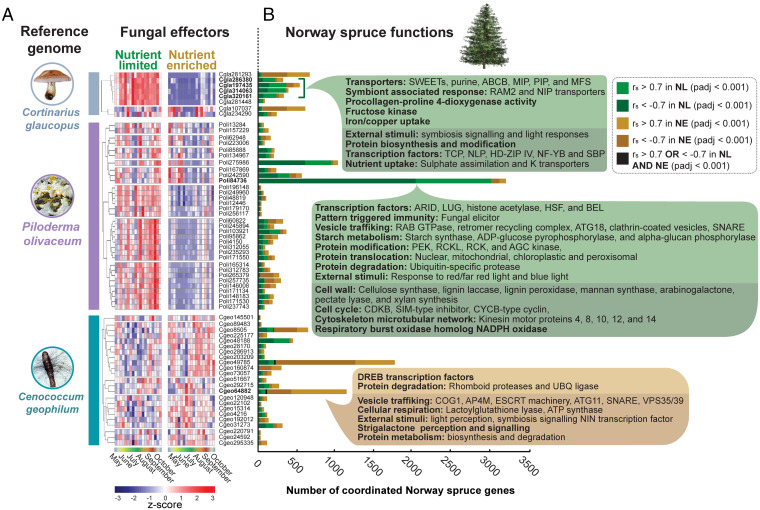
Fig. 5.
Functional analysis of Norway spruce genes displaying positive and negative coordination with predicted fungal effectors. (A) The present set of genes assigned to Co. glaucopus, P. olivaceum, and Ce. geophilum reference genomes were assessed using the fungal effector predictive tool EffectorP (Materials and Methods). The hierarchically clustered transcript abundance of the predicted fungal effectors is displayed for each species. (B) Spearman’s rank correlation was carried out between the expression profiles of the predicted fungal effectors and all Norway spruce transcripts, in both the NL and NE seasonal time courses. The number of Norway spruce genes that satisfied the given Rs (r > 0.7 or < −0.7) and adjusted P thresholds (<0.001) with a given fungal effector are displayed. Genes positively correlated in NL conditions are displayed in light green, negatively correlated in NL are displayed in dark green, positively correlated in NE are displayed in light gold, and negatively correlated in NE are displayed in dark gold. Significant (adjusted P < 0.05) functional enrichments for these gene groups are provided (SI Appendix, Figs. S9–S11). Specific coordinated effectors discussed in Results are marked in bold.