Significance
Photoreceptors utilize sunlight to regulate essential biological processes, and their central machinery domains are potential tools for optogenetics. Among them, the blue light using flavin (BLUF) domain achieves photoswitching functionality via a unique two-way proton-coupled electron transfer (PCET) mechanism. While the forward PCET has been extensively studied with controversial results, the reverse PCET, the key step to trigger the functional conformation switch, is elusive. Here we unambiguously determine the rates and kinetic isotope effects of both forward and reverse PCET in a BLUF domain using ultrafast spectroscopy. When elucidating the nature of the PCET reactions, our work points out that a nearby Trp needs to be mutated. Our findings will facilitate across-species understanding of the photoactivation mechanism in BLUF domains.
Keywords: photoreceptors, proton-coupled electron transfer, proton rocking, stretched dynamics, kinetic isotope effect
Abstract
Proton-coupled electron transfer (PCET) is key to the activation of the blue light using flavin (BLUF) domain photoreceptors. Here, to elucidate the photocycle of the central FMN-Gln-Tyr motif in the BLUF domain of OaPAC, we eliminated the intrinsic interfering W90 in the mutant design. We integrated the stretched exponential function into the target analysis to account for the dynamic heterogeneity arising from the active-site solvation relaxation and the flexible H-bonding network as shown in the molecular dynamics simulation results, facilitating a simplified expression of the kinetics model. We find that, in both the functional wild-type (WT) and the nonfunctional Q48E and Q48A, forward PCET happens in the range of 105 ps to 344 ps, with a kinetic isotope effect (KIE) measured to be ∼1.8 to 2.4, suggesting that the nature of the forward PCET is concerted. Remarkably, only WT proceeds with an ultrafast reverse PCET process (31 ps, KIE = 4.0), characterized by an inverted kinetics of the intermediate FMNH˙. Our results reveal that the reverse PCET is driven by proton transfer via an intervening imidic Gln.
Photoreceptors are a group of functional proteins in nature that utilize light to regulate a wide range of important biological processes such as enzyme activity switching, plant growth and development, animal circadian rhythms, and DNA protection (1). Among them, blue light using flavin (BLUF) domain is one of the smallest photoswitches in nature (2), which functions via an intricate PCET (proton-coupled electron transfer) mechanism. Elucidating the photocycle of the BLUF domain will illuminate how nature utilizes solar energy for biological function via PCET (3–7), which will provide vital guidance for expanding the optogenetic toolboxes (8) as well as for designing new organic synthetic routes (9).
Among all the species, the BLUF domains contain a conservative FMN-Gln-Tyr motif (Fig. 1A). Previous research has suggested that, upon blue-light illumination, the excited-state FMN* receives an electron and a Gln-mediated proton from the nearby Tyr, namely, the forward PCET process. Then, the generated FMNH˙ and Tyr˙ radicals recombine via a more elusive reverse PCET step while the Gln presumably rotates and/or isomerizes, triggering downstream protein conformational changes (10, 11).
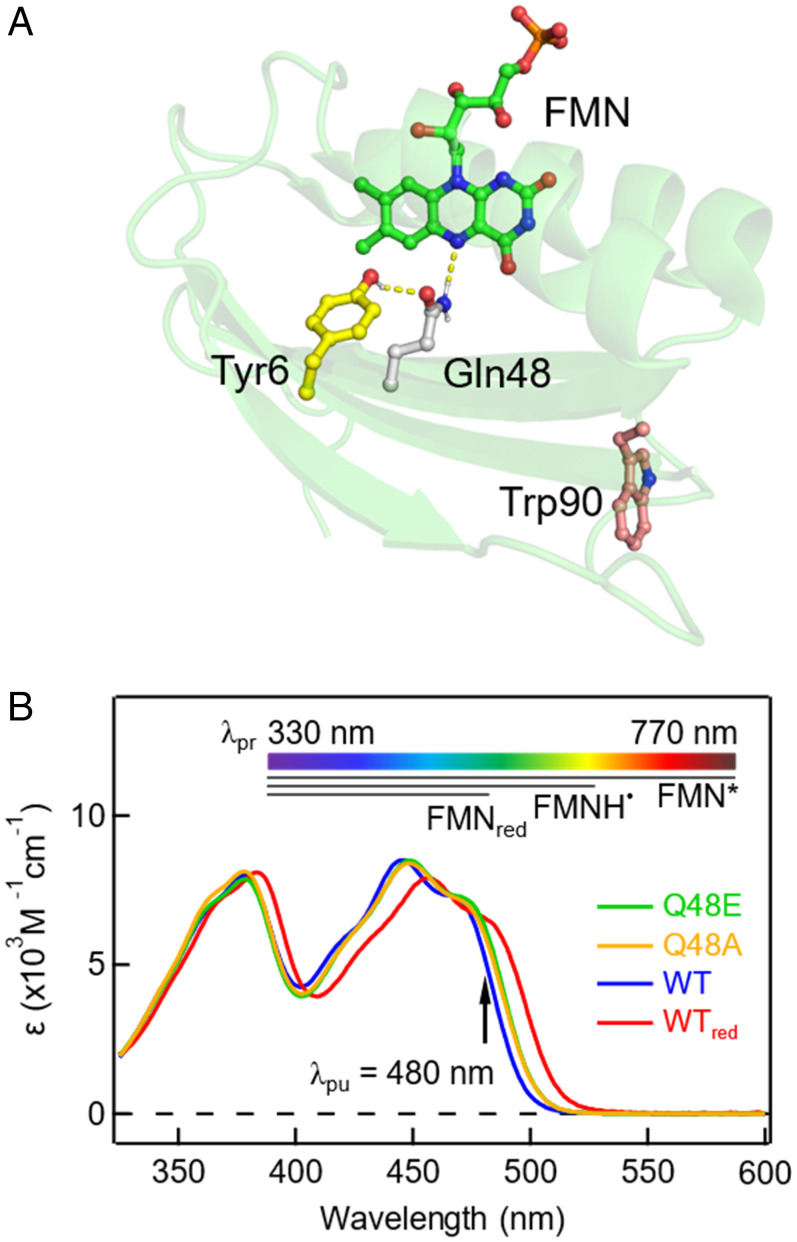
Fig. 1.
Crystal structure and steady-state UV/Vis absorption spectra. (A) Structure of OaPACBLUF (Protein Data Bank ID 4YUT) illustrating the FMN-Gln-Tyr motif in the active site and the nearby W90. (B) Normalized steady-state UV/Vis absorption spectra of OaPACBLUF protein samples where the Q48E, Q48A, WT, and the signaling state WT (referred to as WTred) are indicated with different colors. The pump wavelength is marked by the black arrow. The top color bar denotes the probe range in the TA measurement. The black bars represent the absorption ranges of the excited FMN*, the possible intermediate FMNH˙, and FMNred.
To date, the nature of the forward and reverse PCET remains highly debated. The suggestion that the forward PCET occurs through a sequential eletron transfer/proton transfer (ET/PT) mechanism is based on the detection of FMN˙− intermediates in the BLUF domain of SyPixD, while other species show drastically different photochemistry (10, 12, 13). Intriguingly, recent experimental and theoretical advances on biomimetic systems suggest that, with different ET and PT driving forces, Tyr and Trp can proceed with different PCET mechanisms (3, 7, 14, 15). The progress motivates us to resolve the Tyr-based photochemistry by comparing it with a Trp-substituted one, as well as by considering the interference of a nearby Trp in the BLUF domain (Fig. 1A). On the other hand, the nature of the reverse PCET is unsolved so far, because its kinetics is convoluted with the forward PCET. Experimentally, all BLUF domains have shown a multiexponential decay of the excited-state FMN* with several possible physical origins, which makes the kinetic fitting of the reverse process highly challenging (16, 17), and thus an advanced kinetics analysis strategy with the design of various mutations is required in revealing the nature of the reverse PCET.
In this work, we have resolved the photocycle of the central FMN-Gln-Tyr motif of a BLUF domain from the photoactivated adenylate cyclase (PAC) of the photosynthetic cyanobacterium Oscillatoria acuminata (hereafter referred to as OaPACBLUF) (18, 19). In particular, we have mutated the intrinsic W90 to eliminate a competing ET pathway in all the designed mutants. By comparing the wild type (WT) and its Q48A and Q48E mutants, we have captured an inverted kinetics of the intermediate FMNH˙ in WT featuring a rapid reverse PCET in ∼30 ps. Most significantly, we have developed a global fitting methodology using a stretched exponential function to describe the apparent multiphasic dynamics, allowing the extraction of all the kinetics rates in the photocycle. We performed our experiments and data analysis in both H2O and D2O to obtain the kinetic isotope effect (KIE). Our work bridges the gap between recent advances in the PCET theoretical framework and its applications in biological systems, by revealing the nature of the forward and reverse PCET in the BLUF domain (3, 14, 15).
Results
Unique Mutations and Absorption Properties.
As shown in Fig. 1A, there is an intrinsic W90 in the BLUF domain of OaPAC, which also exists in many other species (20, 21). In most X-ray crystal structures (18, 19, 21), this Trp exhibits a Trpout conformation that is more than 10 Å away from the central motif. However, solution NMR experiments (22) and theoretical calculations (23, 24) have suggested that various Trpin conformations (SI Appendix, Fig. S1) can be populated in buffer solution without the confinement of the crystal lattice. While in the Trpin conformations, the distance between FMN and Trp shortens to ∼3 Å to 4 Å, introducing another light-induced ET pathway (25). Here, we use the single mutant W90F of OaPACBLUF (hereafter referred to as WT for clarity) as a model scaffold to represent the essential FMN-Gln-Tyr motif only.
In order to elucidate the reverse PCET where Q48 plays an important role (26), we have designed two double mutants, Q48E/W90F and Q48A/W90F (referred to as Q48E and Q48A for clarity). The steady-state UV visible (UV/Vis) absorption spectra of the protein samples are illustrated in Fig. 1B. Upon irradiation by a 450-nm diode laser, WT is shown red-shifted by ∼11 nm that persists for 12 s (SI Appendix, Fig. S2), which has been observed in other BLUF domains and is the manifestation of the purported signaling state (WTred, red curve, Fig. 1B) (27, 28). Q48E (green curve, Fig. 1B) and Q48A (orange curve, Fig. 1B) have no absorption change upon blue-light irradiation and feature a slight ∼3-nm red shift with respect to the dark-state WT, which implies changes in the hydrogen-bond environment surrounding FMN and agrees with previous reports on Q mutants in AppA and SyPixD (26, 29). In particular, the small-size Ala substitution leaves space for possible water molecules in the cavity for proton relay. Proton transfer through water chains and/or Glu (as in Q48E) widely exists in biological systems such as in the photosynthetic reaction centers and bacteriorhodopsins (30). By comparing the forward and reverse PCET rates in those mutants, we aim for a deep understanding of the biological relevance of Q48 in the BLUF domain.
Ultrafast Dynamics and Forward PCET.
Broadband femtosecond transient absorption (TA) measurements were carried out on the OaPACBLUF protein samples with the pump wavelength at 480 nm. As shown in Fig. 2 A–C, the original TA contour maps of all three mutants show typical FMN* features at early time delays with positive absorption at above 680 nm, the ground-state bleaching at around 450 nm, and simulated emission signals at around 550 nm (13). Following the initial dynamics of FMN*, the spectra of Q48E show an intermediate species which rises in tens of picoseconds and does not decay within our time window of 7 ns, with a broad absorption at 500 nm to 650 nm and a negative absorption at ∼384 nm, a characteristic of FMNH˙ (SI Appendix, Fig. S3) (31). A similar spectral feature that is attributed to FMNH˙ also appears in tens of picoseconds in Q48A and decays in nanoseconds timescale (Fig. 2B). No discernable signals from FMNH˙ are observed in the original two-dimensional (2D) TA contour of WT yet (Fig. 2C); however, the positive signal at ∼392 nm and 500 nm along with the negative signal at ∼444 nm occurring at longer time delays are indicative of the signaling state (SI Appendix, Fig. S3).
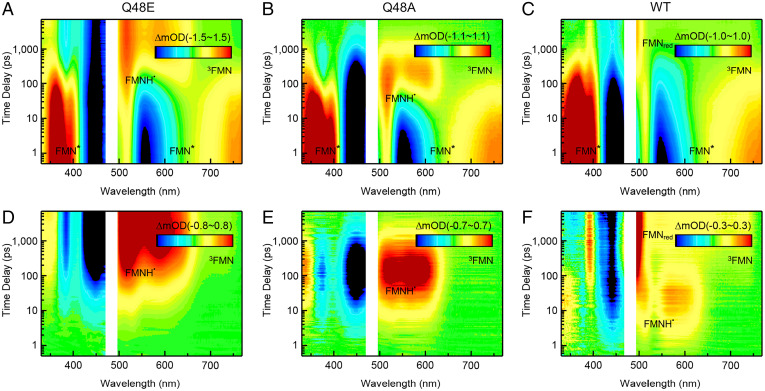
Fig. 2.
TA 2D contour maps. (A–C) Original TA 2D spectra of Q48E, Q48A, and WT in H2O. (D–F) The corresponding TA 2D spectra of Q48E, Q48A, and WT after subtracting the contribution of FMN* from the original data in H2O. The transient species of FMN*, FMNH˙, 3FMN, and FMNred are denoted in the 2D contour maps.
In order to capture all the intermediate species, we subtracted the FMN* contribution from the measured TA data (32, 33) (see SI Appendix, Experimental for a step-by-step tutorial). The subtracted 2D broadband TA spectra of Q48E, Q48A, and WT reveal the appearance of the intermediate species with great clarity, as shown in Fig. 2 D–F. Surprisingly, all the mutants proceed to the PCET reactions upon excitation to form the intermediate FMNH˙ through the transfer of the electron and proton from Y to FMN*, indicating both E and Q can act as a bridge for proton transfer; for the Q48A mutant, the PCET must occur through a bridge of H2O to deliver the proton relay, resulting from a well-defined hydrogen-bond network (see the molecular dynamics [MD] simulation below). Most remarkably, for WT, a weak yet clear intermediate signal appears in several picoseconds and decays in ∼100 ps, with its spectrum resembling a typical FMNH˙ profile before evolving into the signals of FMNred in a sequential way (Fig. 2F). This is unambiguous evidence that, in the photocycle of WT, FMN* also experiences forward PCET and generates FMNH˙, only with drastically fewer populations compared to those in the Q mutants, which can be easily neglected with the overwhelming contribution from FMN*, as shown in Fig. 2C. The observation of FMNH˙ in OaPACBLUF agrees with recent findings by transient infrared (IR) spectroscopy (10, 34). And, unlike those in the Q mutants, only FMNH˙ in WT evolves into FMNred, highlighting the functional significance of Q48. The subtracted 2D TA spectrum of WT also reveals another species growing in nanoseconds, featuring a broad band from 510 nm to 760 nm and a negative band around 357 nm, which is the only product in the photochemistry of ET-inert Y6F/W90F mutant (SI Appendix, Fig. S4), suggesting a pathway of generating 3FMN through intersystem crossing (ISC). In fact, this pathway exists in the Q mutants as well, only masked by the intensive FMNH˙ signals, as resolved by the kinetics model fitting below.
Stretched Behavior, Nonequilibrium Dynamics, and Reverse PCET.
In order to quantify the rates in the PCET processes in the above mutants, we next performed kinetic analysis using the model fitting on selected wavelengths (Fig. 3 A–C). All three samples show a multiphasic decay in their 760-nm kinetics (see SI Appendix, Table S1 for the fitting parameters), which has been observed repeatedly in BLUF domains with debatable physical origins (10, 17). Here, we describe such a multiexponential behavior using a stretched exponential function,, where A, τ, and β represent the amplitude, lifetime, and stretched parameter. The average lifetime can be determined using . The stretched model has been successfully applied in fitting the dynamics of photoinduced ET reactions in photolyases (35), in which case β accounts for the coupling extent between ET and active-site solvation dynamics. On the similar timescales of two processes, in picoseconds, ET could be strongly coupled with the local environment relaxation induced by sudden excitation (36). Here, this approach is applied in the BLUF domain to describe the dynamics of the PCET, and we will show, in later sections, that it greatly reduces the number of fitting parameters and facilitates an elegant kinetic model, a quantification of the KIE, and a physical interpretation of non-single-exponential behavior. Note that all TA matrices are normalized according to the highest absorption at 367 nm to facilitate a direct comparison of the intermediates’ populations.
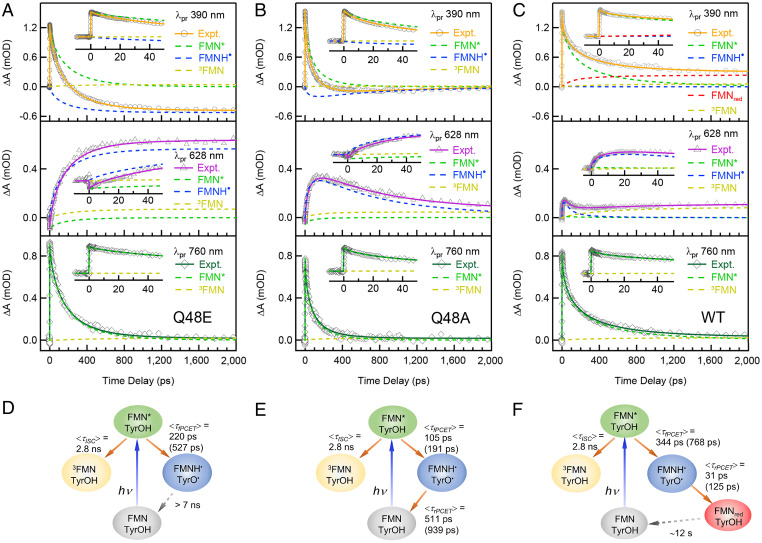
Fig. 3.
Kinetic analysis on TA transients at selected wavelengths. (A–C) TA kinetic traces of Q48E, Q48A, and WT, respectively, in H2O cut from the 2D contour maps at selected wavelengths (760, 628, and 390 nm), with the decomposed dynamics of the reactant (FMN*), the intermediate (FMNH˙), and the products (FMNred and 3FMN). Early-time TA transients and decomposed dynamics up to 50 ps are shown in the Insets of A–C. Raw data are represented with symbols, and the fitted kinetics are represented with straight lines. The kinetics of the given species are shown in colored dash lines. The proposed kinetic models and the fitted rates in H2O and D2O (in parentheses) for Q48E, Q48A, and WT are shown in D, E, and F, respectively.
Specifically, at 760 nm, the absorption transients in all three mutants (Fig. 3) arise mostly from FMN*, with some minor contributions from 3FMN. The transients can be fitted using a parallel kinetics model (Fig. 3 D–F) with a fixed 2.8-ns (obtained from ET-inert Y6F/W90F mutant; SI Appendix, Fig. S4) ISC branch and a forward PCET pathway, with the average rates of 105 ps to 344 ps and a β value of 0.5 to 0.71, as shown in Table 1. The transients at 628 nm are dominated by the kinetics of FMNH˙, with some minor contributions from FMN* and 3FMN. In Q48E, FMNH˙ rises with the same rate as the FMN* decay (∼220 ps, β = 0.71) and remains constant within the time window, so it is consistent with a model shown in Fig. 3D. For Q48A, the kinetics of FMNH˙ at 628 nm shows a rise (∼105 ps, β = 0.63) that matches the FMN* decay and then a decay with an average rate of 511 ps, β = 0.57 (Table 1), agreeing with the photocycle shown in Fig. 3E. Most importantly, for WT, after subtracting the contribution from the FMN*, the FMNH˙ transient at 628 nm shows an ultrafast rise in 31 ps (β = 0.92) and a decay of the same rate as the decay of the FMN* dynamics at 760 nm (344 ps, β = 0.5) (Table 1). Together with the lowest population of FMNH˙ in WT among the three mutants, this transient has a characteristic of so-called inverted kinetics (see SI Appendix, Fig. S5 for an illustration of a simple sequential model), where the rise of the kinetics shows the reverse PCET, and the decay actually is the formation of FMNH˙ from the forward PCET, as shown in the model in Fig. 3F. At 390 nm, the two Q mutants can be fitted with a sum of the kinetics of FMN*, FMNH˙, and 3FMN, while only WT requires an extra FMNred component which rises and remains constant within our time window, consistent with the proposed model (Fig. 3F).
Table 1.
The fitting results of OaPACBLUF mutants based on target analysis using the kinetic models
| Mutants | Parameters* | H2O | D2O | KIE† |
|---|---|---|---|---|
| Q48E | τfPCET | 176 | 392 | |
| βfPCET | 0.71 | 0.66 | ||
| <τfPCET> | 220 | 527 | 2.4 | |
| Q48A | τfPCET | 74 | 127 | |
| βfPCET | 0.63 | 0.60 | ||
| <τfPCET> | 105 | 191 | 1.8 | |
| τrPCET | 317 | 567 | ||
| βrPCET | 0.57 | 0.56 | ||
| <τrPCET> | 511 | 939 | 1.8 | |
| WT | τfPCET | 172 | 231 | |
| βfPCET | 0.50 | 0.40 | ||
| <τfPCET> | 344 | 768 | 2.2 | |
| τrPCET | 30 | 87 | ||
| βrPCET | 0.92 | 0.62 | ||
| <τrPCET> | 31 | 125 | 4.0 |
*All average lifetimes <τ> (ps) are calculated using in picoseconds, with the lifetimes τ (picoseconds), and the stretched parameters β, and are used as reported lifetimes throughout the manuscript. fPCET in the subscript is short for forward PCET and rPCET reverse PCET.
†All KIE values are calculated using kH/kD = <τ>D/<τ>H.
Difference Spectra, Transient Intermediates, and Isotope Effects.
In order to obtain the KIE (KIE = kH/kD) of the forward and reverse PCET in OaPACBLUF mutants, we also conducted TA experiments in D2O solution. The 2D TA matrices and the transients at selected wavelengths for the three mutants in D2O have been analyzed using the same approach as above (SI Appendix, Figs. S6 and S7). According to the proposed model shown in Fig. 3 D–F, we decomposed the TA matrices in H2O and D2O into species-associated difference spectra (SADS) and respective kinetic traces by fitting all of the wavelengths with the kinetics model (see SI Appendix, Experimental for a step-by-step tutorial). Such analysis not only allows us to check the correctness of the proposed kinetics model but also facilitates the capture of other possible intermediates by scrutinizing the features of the SADS. The target analysis results of WT are shown in Fig. 4, and those for the Q mutants are shown in SI Appendix, Figs. S8 and S9. Remarkably, the corresponding SADS of the transient intermediates in H2O and D2O match very well with each other, indicating a successful matrix decomposition. The green SADS (Fig. 4 A and C) show typical characteristics of FMN* for flavoproteins (SI Appendix, Fig. S3) (37). The blue SADS component has positive broad absorptions in 500 nm to 650 nm and a negative peak at 384 nm that are attributed to FMNH˙ (SI Appendix, Fig. S3) (31). Most interestingly, in comparison with the FMNH˙/W˙ SADS of our recently reported Y6W/W90F mutant which is a W analog of WT (37), we discover clear spectral signatures of YO˙ (38, 39) around 410 nm to 420 nm for WT, which is also observed in the Q48A and Q48E mutants (SI Appendix, Fig. S10). This suggests that YO˙ appears along with FMNH˙, which is evidence that the proton relays in a Grotthuss-type way. The red SADS are consistent with the steady-state light minus dark spectrum of WT (SI Appendix, Fig. S3), and the orange SADS are the triplet 3FMN spectrum obtained from the PCET-inert Y6F/W90F mutant (SI Appendix, Fig. S4).
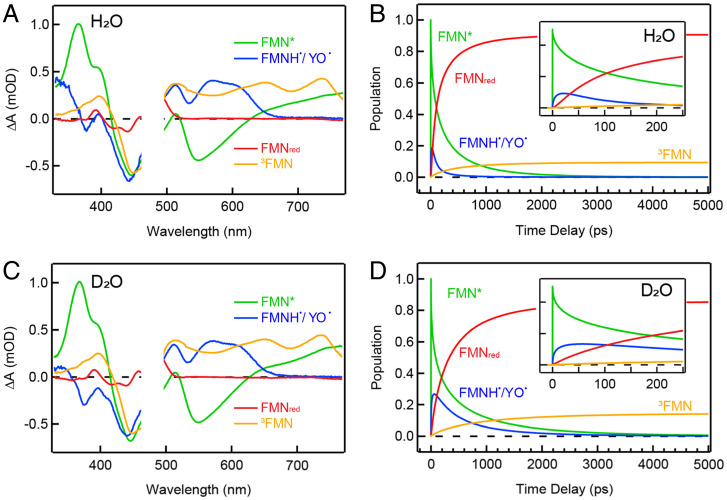
Fig. 4.
Target analysis results. SADS (A and C) and corresponding kinetic traces (B and D) of WT in H2O and D2O fitted with the proposed kinetic model as shown in Fig. 3F. Early-time kinetic traces up to 250 ps are shown in the Insets of B and D. In the kinetic trace diagrams, transient species of FMN*, FMNH˙/YO˙, FMNred, and 3FMN are indicated with different colors.
Important KIE information on the forward and reverse PCET can be obtained from the fitted parameters (Table 1) using the above kinetics model in Fig. 3, and is manifested in the decomposed kinetic traces from the target analysis. Notably, for WT (Fig. 4), the forward PCET (344 ps in H2O and 768 ps in D2O) is much slower than the reverse (31 ps in H2O and 125 ps in D2O), resulting in an inverted kinetics of the FMNH˙/YO˙ traces in both cases (Fig. 4). The population of FMNH˙/YO˙ clearly accumulates more in D2O than in H2O, due to the more significant KIE of reverse PCET (4.0) compared to the forward (2.2). Interestingly, the KIEs of the forward PCET for Q48A and Q48E are 1.8 and 2.4, respectively, similar to that for WT. Together with the forward PCET rates ranging from 105 ps to 344 ps on the same order of magnitude in H2O, this observation suggests that the forward PCET shares some similarity among the three mutants. On the other hand, the reverse PCET in Q48A (511 ps in H2O) and Q48E (the population of FMNH˙/YO˙ remains constant within our time window) happens much more slowly than that in WT (31 ps in H2O). Besides, while the KIE in the reverse pathway in Q48A is the same as that in the forward (∼1.8), in WT, the reverse PCET shows a more significant KIE (∼4) than the forward. It is noteworthy that both forward and reverse PCET consist of multiple elementary steps, including the electron transfer and a two-step proton transfer, with some of them occurring in a concerted fashion. Thus, the KIE values acquired here cover the entire pathway.
Hydrogen-Bond Network and Dynamic Heterogeneity.
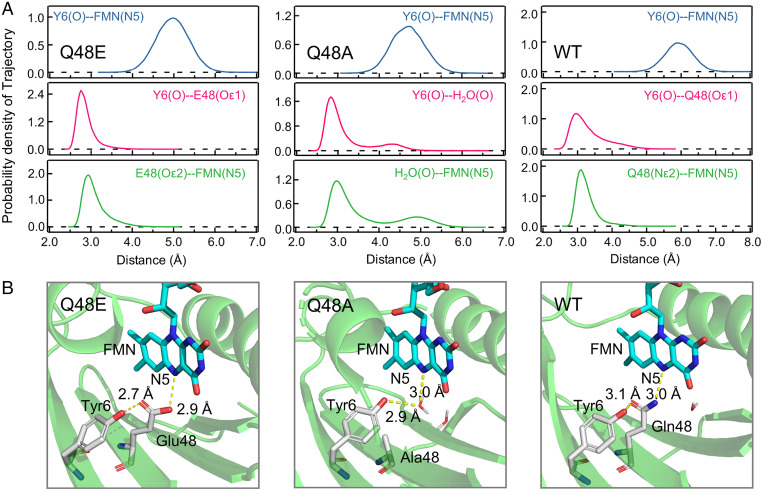
Proton transfer is very sensitive to the donor–acceptor distance and happens only when the donor and acceptor are within H-bonding distance. Thus, we carried out an MD simulation to map the H-bonding network in the active site. The distance between the Tyr phenol O and FMN N5 is much longer than a normal H-bonding distance in all mutants (Fig. 5), which suggests that the proton on Tyr cannot directly transfer to FMN. E48 is reportedly protonated in the Q63E mutant of the AppA BLUF domain (counterpart of the Q48E mutant in this work) evidenced by the 1,724 cm−1 side chain absorption in the transient IR spectra (40). Thus, our MD simulations of Q48E consist of a protonated Glu as an initial configuration, and we observed from the trajectories that there is a stable H-bond existing between the Tyr phenol O and the Glu side chain carbonyl Oε1 (2.6 Å to 3.2 Å), and transient H-bonds can be formed between protonated Glu side chain hydroxyl Oε2 and FMN N5 (2.8 Å to 3.6 Å) (Fig. 5B). In Q48A, MD simulation results show that there is a bound water molecule in the cavity most of the time within the H-bonding distance to both Tyr phenol O (2.6 Å to 3.4 Å) and FMN N5 (2.8 Å to 3.5 Å), with a minor population H-bonded to the C4=O of FMN (SI Appendix, Fig. S11). Significantly, here we found that a single trapped water molecule in a triad FMN-H2O-Tyr, which proceeds to a forward and then reverse PCET with a two-step proton relay in each process, acts a key role in bridging both the donor and acceptor within H-bond distances. This finding may also explain the PCET reactions observed in the Q50A mutant of SyPixD (26). The MD simulation results of WT show a geometry that consists of transient H-bonds between the carbonyl Oε1 on the Gln side chain and the Tyr phenol O (2.8 Å to 3.5 Å), as well as between the Gln side chain amino group Nε2 and FMN N5 (2.9 Å to 3.5 Å). This H-bonding network is similar to the proton relay conformation proposed by the Hammes–Schiffer group for the PixD BLUF domain (24), which presumably can effectively relay a Gln-mediated two-step proton transfer. In all, our MD simulations show that Q48E, Q48A, and WT position a protonated Glu, a structural water, and an oriented Gln, respectively, as a PT bridge. Interestingly, our MD simulation results show that Q48E forms the strongest H-bonds with the shortest distances between heavy atoms, while the H-bonds in WT are weakest, with the largest distance fluctuations, indicating the flexibility of the Gln side chain and dynamic structural heterogeneity in WT. We will elaborate, in the discussion below, that this observation can be correlated to the fitting results that WT has the smallest β value for the forward PCET, while Q48E has the largest, which means the stretched parameter β also reflects how the H-bond fluctuations affect the PT-coupled ET. Besides the H-bond network dynamic heterogeneity, we examined the local hydration and found that, although the FMN-bridge-Tyr sites do not contain dynamic water (within 5-Å distances to FMN N5), there are ∼60 water molecules within 10-Å distances of the isoalloxazine ring of FMN in all mutants, mostly sandwiched between the isoalloxazine ring and the α-helices (SI Appendix, Fig. S12). There are three typical solvation timescales in flavoproteins as measured by ultrafast up-conversion spectroscopy (41). The first ultrafast motion is within several picoseconds, manifesting the water local relaxation. The second hydration timescale is on tens of picoseconds and is originated from the local water-network rearrangement, and the third one (hundreds of picoseconds) reflects the coupled water–protein fluctuation. These three types of solvation dynamics can couple with the PCET processes as observed in other flavoproteins (35, 36).
Fig. 5.
MD simulations. (A) Distributions of the corresponding simulated distances between possible H-bond donors and acceptors of Q48E, Q48A, and WT calculated from the 200-ns MD simulation trajectories. (B) The representative snapshots to illustrate the hydrogen-bond network in Q48E, Q48A, and WT.
Discussion
We aim to reveal the nature of the forward and reverse PCET in the central motif of the OaPAC BLUF domain by summarizing the obtained kinetic rates and KIE values in our work, as well as by comparing them with our recently resolved dynamics of Y6W/W90F, where Y6 is substituted into Trp (Fig. 6) (37). The forward PCET is known to start with the light-induced ET with two possibilities, namely, sequential ET/PT or CEPT (concerted electron–proton transfer) process. First, we did not capture the FMN˙− signals in all three Tyr-based mutants, yet, in Y6W/W90F, we clearly resolved the intermediate species spectra of FMN˙− and WH˙+ (SI Appendix, Fig. S13). Second, all three Tyr-based mutants show notable KIEs (1.8 to 2.4; Table 1) for the 760-nm decay, while Y6W/W90F exhibits almost no KIE (1.1) at 760 nm. With the information above, it is straightforward to conclude that Trp proceeds to sequential ET/PT while Tyr probably undergoes a CEPT mechanism. The reported redox potentials of Y˙+/Y ≈ +1.8 eV and W˙+/W ≈ +1.15 eV (42) indicate that Trp has a larger ET driving force by 0.65 eV. The pKa of Y˙+ is around −2, while, for W˙+, it is ∼4, suggesting that Y˙+ has a larger PT driving force (7). Thus, the discrepancy in the Trp- and Tyr-based PCET in our work agrees remarkably with the trend observed in the biomimetic systems previously (14, 15), and the work here shows an elegant demonstration of the PCET theory in a natural biological system. In addition, when we mutate Q48 into Leu with a bulky hydrophobic side chain to remove any possible proton donor, the ET of Y and FMN is completely abolished, with the fluorescence emission and the triplet 3FMN formation as the main deactivation channels in nanoseconds (SI Appendix, Fig. S14), further corroborating that the initial reaction is CEPT in WT. Here, we emphasize again the necessity to exclude W90 in resolving the photochemistry, with the knowledge that Trp and Tyr may proceed via different PCET mechanisms. Indeed, we show, in SI Appendix, Fig. S15, the FMN* subtracted 2D contour map of the native WT with the retaining of W90, which clearly displays the intermediate FMN˙− and WH˙+ species. Last, we explore the physical origin of the multiphasic kinetics, using a stretched parameter β in the range of 0.4 to 0.71 to better reflect the dynamic heterogeneity in the forward PCET processes of our mutants. Y6W/W90F shows a single-exponential decay of the FMN* in 2.2 ps (37), with a large β value close to one. This observation indicates that the coupling of ET with the solvation dynamics is minor. For the Tyr-based proteins with removal of W90 (WT, Q48A, and Q48E), we observed a much longer PCET dynamics in 105 ps to 344 ps with β ranging from 0.5 to 0.71 in H2O, a significant stretched behavior, indicating a strong coupling with environment relaxations until hundreds of picoseconds, leading to a small β. More importantly, the β value for WT (0.5) is unprecedentedly small, which cannot be explained by the solvation relaxation only. This observation suggests that β is also a strong indicator for the coupling of ET with the PT, the extent of which is determined by the dynamic structural heterogeneity of the H-bond network which has been observed in the MD simulations, due to the high flexibility of the Gln side chain and a concerted forward PCET, not sequential. The flexible Gln side chain introduces a dynamic heterogeneity in the proton donor–acceptor distances in the PCET theory (3, 15) and may result in a highly stretched dynamic behavior. Notably, with the ultrafast reverse PCET in 31 ps, the β value becomes larger, 0.92, a less coupled process with local solvation, which is on picosecond timescales (41). Our simulation is performed on the ground state, yet, after the photoinduced ET, the FMN-Gln-Tyr motif might become more compact due to the electrostatic interaction of the charge-separated pair, which might reduce the H-bond flexibility and heterogeneity of the proton donor–acceptor distances, resulting in a larger β value. Our work shows that it is crucial to describe the multiphasic dynamics in a PCET reaction using a stretched function. This approach not only is key to the accurate determination of the rates and KIE values for the elusive reverse PCET process in the BLUF domain which are previously unattainable, but it also provides important insights into the dynamics or nonergodicity of the PCET process in a general sense (43, 44).
Fig. 6.
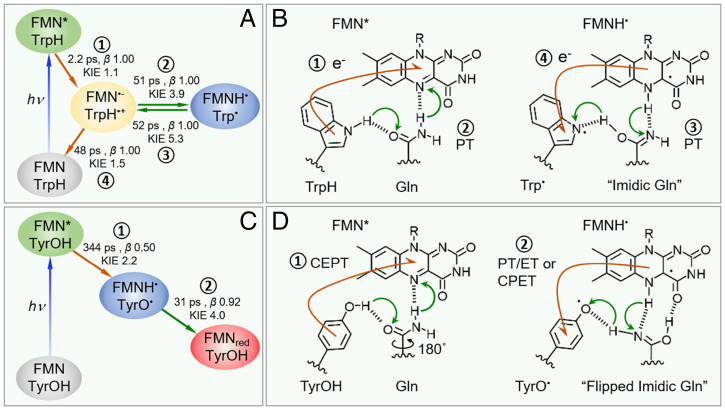
Proposed model for OaPACBLUF domain photochemistry. Kinetics model with fitted parameters from ultrafast measurements (A) and PCET mechanism (B) of Y6W/W90F mutant (37), and the mechanism proceeds with sequential forward ET (1) and forward PT (2) after photoexcitation of FMN followed with sequential reverse PT (3) and reverse ET (4) where the respective rates, β, and KIE are given in A. (C and D) The kinetics model and PCET mechanisms for WT. In WT, the mechanism proceeds with forward CEPT (1) followed by either reverse sequential PT/ET or reverse CPET (2) where the corresponding rates, β, and KIE are given in C. Flows of the electrons are denoted by orange arrows, and protons are denoted by green arrows, in B and D.
Next, we discuss the nature of the reverse PCET. First, only WT, rather than the nonfunctional Q48A and Q48E, experiences an ultrafast reverse PCET (∼31 ps). Interestingly, we also found that Y6W/W90F can rock a proton back in an ultrafast fashion (∼52 ps) (37), a timescale similar to that in WT. Second, WT has a significant KIE effect (4.0), similar to that observed in Y6W/W90F (5.3). Although, in Y6W/W90F, the reverse PCET via a sequential PT/ET is clearly resolved with the intermediates FMN˙− and WH˙+ species captured (37), WT here could be either sequential PT/ET where PT is the rate-determining step or CPET (concerted proton–electron transfer), because the key FMN˙−/Tyr˙+ intermediates are not obviously observed. Nonetheless, from the significant KIE (4.0), we can conclude that the reverse PCET is driven by the proton transfer via Gln, not ET/PT. This is direct experimental evidence that proton rocking exists in the WT BLUF domain, after the Batista and Malvankar groups proposed this notion based on the similarity of the Tyr-Gln geometry in the BLUF domain to a conducting amyloid (45). The large discrepancy of the three mutants in the reverse pathway suggests that the pKa values of the bridge on both the proton acceptor and donor moieties determine the rates or the occurrence of the reverse processes. Further endeavor is required to dissect the two proton-transfer steps in each relay in order to reveal the rate and energetics underlying the proton rocking process.
We then ask, why WT can induce a spectral shift while Y6W/W90F cannot if both undergo an ultrafast reverse PCET? As shown in Fig. 6, after the forward PCET, Gln in both cases lands in an imidic acid form as proposed before (46–48). However, before the reverse PCET, only in WT, not Y6W/W90F, the imidic Gln experiences an extra rotation and forms a new H-bond with the C4=O of FMN which has been suggested by previous IR work (16, 49) and calculations (46–48). From our MD simulations (SI Appendix, Fig. S16), the average distance between Tyr O and FMN N5 in WT is 5.9 Å, while Trp N and FMN N5 in Y6W/W90F is 6.9 Å, and the latter is too far to be co-H-bonded with a single N atom, as in the case of WT (Fig. 6). Thus, for the reverse PCET, the actual proton rocking pathways are likely different for WT and Y6W/W90F (Fig. 6), highlighting the synergetic role of Tyr and Gln in realizing the biological function. Interestingly, we are aware that, in this model, the forward PCET processes for the dark state and the light state involve the amine C=O and the imidic N of Gln as the proton acceptor with drastically different pKa. If we assume both are concerted PCET, it can lead to a large difference in the energetic term in the PCET theory (3, 15) which explains the one to two orders of magnitude discrepancy in the PCET rates for the dark and light states as reported (17).
We noticed that recent X-ray crystallographic work on the dark- and light-state full-length OaPAC has suggested an alternative photocycle featuring subtle side chain movements instead of the Q48 flipping (18), which coincided with the photoactivation pathway deduced from theoretical calculations (50) and a recent ultrafast experiment (33). This model is also largely coherent with our kinetic results, with the only inconsistency being that it shares a reverse PCET pathway similar to that in the FMN-H2O-Tyr triad and cannot be used to explain the 16-fold discrepancy in the rates of the reverse PCET for WT (31 ps) and Q48A (511 ps) (SI Appendix, Fig. S17). Nonetheless, this alternative model is quite insightful, and further temperature-dependent experiments are required to investigate the purported Cγ–Cδ rotation of Q48 during the photocycle.
Although our results with OaPACBLUF do not necessarily expand to other BLUF species, our strategy in mutant design and target analysis can be generalized and potentially can reveal an across-species mechanism. For example, the previous work on SyPixD suggested a forward sequential ET/PT mechanism due to the detection of the FMN˙− intermediate species (12, 13). However, the peculiarity of SyPixD has been noticed because only neutral intermediates have been observed in other species (10, 32). We are aware that, in the X-ray crystal structures of SyPixD, 2 out of 10 subunits adapted Trpin conformations (21). Moreover, calculations have suggested that more Trpin conformations may be populated in solution (24). Together with our results in OaPAC (SI Appendix, Fig. S15), the possibility that the observed FMN˙− in SyPixD is actually from an interfering FMN-Trp ET pathway needs to be reconsidered, which might be the reason why the simulated dark-state PCET timescale is much faster than the experimentally assigned one (51). We also need to be cautious about using the FMN˙− intermediate species as evidence for concerted PCET. If the forward ET is only concerted with the first PT step, but not the second, there is also a chance to observe minimal amounts of FMN˙− intermediate, depending on the rate of each step.
In the current work, we elucidated the early photochemistry of the OaPACBLUF WT and two Q mutants (Q48A and Q48E) using ultrafast spectroscopy. We have excluded the interference of W90, to avoid the complexity of ET pathways. We found that the nature of the forward PCET in WT (344 ps in H2O, KIE 2.2) is likely CEPT. One of the key approaches is to untangle the early photochemistry using a stretched exponential function to describe the multiphasic dynamics. Our MD simulation results show that the Gln H-bonds in FMN-Gln-Tyr motif are flexible, accounting for the relatively small β of 0.5 in the forward PCET of WT. Significantly, different from Q48A and Q48E, the reverse PCET process in WT is ultrafast (31 ps) and less stretched (β = 0.92), resulting in the inverted kinetics behavior in the FMNH˙/YO˙ transients. Together with the KIE of 4.0 in the reverse PCET of WT, our results provide direct experimental evidence that the reverse PCET process is driven by the proton transfer via an imidic Gln. Thus, our findings can facilitate the understanding of the early photochemistry across the BLUF domain species and provide important insights into the nonequilibrium dynamics of the PCET processes in biology.
Materials and Methods
Detailed information on materials and methods is provided in SI Appendix, Experimental, including plasmid construction and primer design, protein expression and purification, steady-state spectroscopy, TA spectroscopy, a step-by-step tutorial on data analysis, and MD simulations.
Protein Preparation.
All of the studied OaPACBLUF mutants were expressed and purified with a standard laboratory method as elaborated in SI Appendix, Experimental. The purified protein was stored at −80 °C after being quickly frozen with liquid nitrogen. All of the protein samples were diluted to 200 μM to 300 μM by flowing through a mini desalting column to get rid of unbound FMN molecules before TA experiments.
TA Spectroscopy.
The TA spectra were measured by a commercial TA spectrometer (Helios Fire, Ultrafast Systems). All the protein samples were pumped at 480 nm. The pump power was attenuated to 0.1 mW. The polarization between the pump and the probe pulses was set to be the magic angle (54.7°). The samples were continuously stirred by a magnetic stir bar in the 2-mm-path-length cuvette to ensure the refreshing of the sample for each pulse. More experimental details can be found in SI Appendix, Experimental.
Data Analysis.
The TA data were first globally analyzed using a sequential model to obtain the spectrum of FMN*. The decay of FMN* was obtained by fitting the 760-nm kinetics. Then the FMN* component was subtracted from the raw TA data matrix to show the evolution of transient intermediates such as FMNH˙ and FMNred. A stretched parameter β was involved in the rate equation to describe the multiphasic kinetics. The target analyses were conducted using a script written in MATLAB to extract the SADS spectra. A detailed step-by-step tutorial on data analysis is provided in SI Appendix, Experimental.
Supplementary Material
Acknowledgments
B.D. thanks Dr. Yangyi Lu for stimulating discussions. We thank Prof. Juntao Ye for discussing organic reaction mechanisms involved in this work. We thank Dr. Faming Lu for maintaining the laser facility. Funding support to B.D. from the National Natural Science Foundation of China (Grants 31971233 and 21903052) and the National Key Research and Development Program of China (Grant 2020YFA0509700) are acknowledged.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2203996119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Seong J., Lin M. Z., Optobiochemistry: Genetically encoded control of protein activity by light. Annu. Rev. Biochem. 90, 475–501 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zoltowski B. D., Gardner K. H., Tripping the light fantastic: Blue-light photoreceptors as examples of environmentally modulated protein-protein interactions. Biochemistry 50, 4–16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hammes-Schiffer S., Stuchebrukhov A. A., Theory of coupled electron and proton transfer reactions. Chem. Rev. 110, 6939–6960 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reece S. Y., Nocera D. G., Proton-coupled electron transfer in biology: Results from synergistic studies in natural and model systems. Annu. Rev. Biochem. 78, 673–699 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dempsey J. L., Winkler J. R., Gray H. B., Proton-coupled electron flow in protein redox machines. Chem. Rev. 110, 7024–7039 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinberg D. R., et al. , Proton-coupled electron transfer. Chem. Rev. 112, 4016–4093 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Migliore A., Polizzi N. F., Therien M. J., Beratan D. N., Biochemistry and theory of proton-coupled electron transfer. Chem. Rev. 114, 3381–3465 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fenno L., Yizhar O., Deisseroth K., The development and application of optogenetics. Annu. Rev. Neurosci. 34, 389–412 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin N. Y., Ryss J. M., Zhang X., Miller S. J., Knowles R. R., Light-driven deracemization enabled by excited-state electron transfer. Science 366, 364–369 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lukacs A., Tonge P. J., Meech S. R., Photophysics of the blue light using flavin domain. Acc. Chem. Res. 55, 402–414 (2022). [DOI] [PubMed] [Google Scholar]
- 11.Fujisawa T., Masuda S., Light-induced chromophore and protein responses and mechanical signal transduction of BLUF proteins. Biophys. Rev. 10, 327–337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonetti C., et al. , Hydrogen bond switching among flavin and amino acid side chains in the BLUF photoreceptor observed by ultrafast infrared spectroscopy. Biophys. J. 95, 4790–4802 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gauden M., et al. , Hydrogen-bond switching through a radical pair mechanism in a flavin-binding photoreceptor. Proc. Natl. Acad. Sci. U.S.A. 103, 10895–10900 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agarwal R. G., et al. , Free energies of proton-coupled electron transfer reagents and their applications. Chem. Rev. 122, 1–49 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tyburski R., Liu T., Glover S. D., Hammarström L., Proton-coupled electron transfer guidelines, fair and square. J. Am. Chem. Soc. 143, 560–576 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukacs A., et al. , BLUF domain function does not require a metastable radical intermediate state. J. Am. Chem. Soc. 136, 4605–4615 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kennis J. T. M., Mathes T., Molecular eyes: Proteins that transform light into biological information. Interface Focus 3, 20130005 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohki M., et al. , Molecular mechanism of photoactivation of a light-regulated adenylate cyclase. Proc. Natl. Acad. Sci. U.S.A. 114, 8562–8567 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohki M., et al. , Structural insight into photoactivation of an adenylate cyclase from a photosynthetic cyanobacterium. Proc. Natl. Acad. Sci. U.S.A. 113, 6659–6664 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson S., et al. , Structure of a novel photoreceptor, the BLUF domain of AppA from Rhodobacter sphaeroides. Biochemistry 44, 7998–8005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan H., et al. , Crystal structures of the Synechocystis photoreceptor Slr1694 reveal distinct structural states related to signaling. Biochemistry 45, 12687–12694 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grinstead J. S., et al. , The solution structure of the AppA BLUF domain: Insight into the mechanism of light-induced signaling. ChemBioChem 7, 187–193 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Goyal P., Hammes-Schiffer S., Role of active site conformational changes in photocycle activation of the AppA BLUF photoreceptor. Proc. Natl. Acad. Sci. U.S.A. 114, 1480–1485 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goings J. J., Reinhardt C. R., Hammes-Schiffer S., Propensity for proton relay and electrostatic impact of protein reorganization in Slr1694 BLUF photoreceptor. J. Am. Chem. Soc. 140, 15241–15251 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Laan W., et al. , On the mechanism of activation of the BLUF domain of AppA. Biochemistry 45, 51–60 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Fudim R., et al. , Photoinduced formation of flavin radicals in BLUF domains lacking the central glutamine. FEBS J. 282, 3161–3174 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Hasegawa K., Masuda S., Ono T. A., Structural intermediate in the photocycle of a BLUF (sensor of blue light using FAD) protein Slr1694 in a cyanobacterium Synechocystis sp. PCC6803. Biochemistry 43, 14979–14986 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Masuda S., Bauer C. E., AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell 110, 613–623 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Dragnea V., Arunkumar A. I., Lee C. W., Giedroc D. P., Bauer C. E., A Q63E Rhodobacter sphaeroides AppA BLUF domain mutant is locked in a pseudo-light-excited signaling state. Biochemistry 49, 10682–10690 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugo Y., Saito K., Ishikita H., Mechanism of the formation of proton transfer pathways in photosynthetic reaction centers. Proc. Natl. Acad. Sci. U.S.A. 118, e2103203118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang M., Wang L., Zhong D., Photolyase: Dynamics and electron-transfer mechanisms of DNA repair. Arch. Biochem. Biophys. 632, 158–174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujisawa T., Takeuchi S., Masuda S., Tahara T., Signaling-state formation mechanism of a BLUF protein PapB from the purple bacterium Rhodopseudomonas palustris studied by femtosecond time-resolved absorption spectroscopy. J. Phys. Chem. B 118, 14761–14773 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Fujisawa T., Masuda S., Takeuchi S., Tahara T., Femtosecond time-resolved absorption study of signaling state of a BLUF protein PixD from the cyanobacterium Synechocystis: Hydrogen-bond rearrangement completes during forward proton-coupled electron transfer. J. Phys. Chem. B 125, 12154–12165 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Lukacs A., et al. , Radical formation in the photoactivated adenylate cyclase OaPAC revealed by ultrafast spectroscopy. Biophys. J. 118, 608a (2020). [Google Scholar]
- 35.Li J., et al. , Dynamics and mechanism of repair of ultraviolet-induced (6-4) photoproduct by photolyase. Nature 466, 887–890 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu Y., Kundu M., Zhong D., Effects of nonequilibrium fluctuations on ultrafast short-range electron transfer dynamics. Nat. Commun. 11, 2822 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang X.- W., et al. , Direct observation of ultrafast proton rocking in the BLUF domain. Angew. Chem. Int. Ed. Engl. 61, e202114423 (2022). [DOI] [PubMed] [Google Scholar]
- 38.Oldemeyer S., et al. , Essential role of an unusually long-lived tyrosyl radical in the response to red light of the animal-like cryptochrome aCRY. J. Biol. Chem. 291, 14062–14071 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glover S. D., et al. , Photochemical tyrosine oxidation in the structurally well-defined α3Y protein: Proton-coupled electron transfer and a long-lived tyrosine radical. J. Am. Chem. Soc. 136, 14039–14051 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lukacs A., et al. , Photoexcitation of the blue light using FAD photoreceptor AppA results in ultrafast changes to the protein matrix. J. Am. Chem. Soc. 133, 16893–16900 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Chang C.- W., et al. , Mapping solvation dynamics at the function site of flavodoxin in three redox states. J. Am. Chem. Soc. 132, 12741–12747 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishikita H., Light-induced hydrogen bonding pattern and driving force of electron transfer in AppA BLUF domain photoreceptor. J. Biol. Chem. 283, 30618–30623 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matyushov D. V., Protein electron transfer: Is biology (thermo)dynamic? J. Phys. Condens. Matter 27, 473001 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Soudackov A., Hammes-Schiffer S., Multistate continuum theory for multiple charge transfer reactions in solution. J. Chem. Phys. 111, 4672–4687 (1999). [Google Scholar]
- 45.Shipps C., et al. , Intrinsic electronic conductivity of individual atomically resolved amyloid crystals reveals micrometer-long hole hopping via tyrosines. Proc. Natl. Acad. Sci. U.S.A. 118, e2014139118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goings J. J., Li P., Zhu Q., Hammes-Schiffer S., Formation of an unusual glutamine tautomer in a blue light using flavin photocycle characterizes the light-adapted state. Proc. Natl. Acad. Sci. U.S.A. 117, 26626–26632 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Domratcheva T., Hartmann E., Schlichting I., Kottke T., Evidence for tautomerisation of glutamine in BLUF blue light receptors by vibrational spectroscopy and computational chemistry. Sci. Rep. 6, 22669 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Domratcheva T., Grigorenko B. L., Schlichting I., Nemukhin A. V., Molecular models predict light-induced glutamine tautomerization in BLUF photoreceptors. Biophys. J. 94, 3872–3879 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iwata T., et al. , Hydrogen bonding environments in the photocycle process around the flavin chromophore of the AppA-BLUF domain. J. Am. Chem. Soc. 140, 11982–11991 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Sadeghian K., Bocola M., Schütz M., A conclusive mechanism of the photoinduced reaction cascade in blue light using flavin photoreceptors. J. Am. Chem. Soc. 130, 12501–12513 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Goings J. J., Hammes-Schiffer S., Early photocycle of Slr1694 blue-light using flavin photoreceptor unraveled through adiabatic excited-state quantum mechanical/molecular mechanical dynamics. J. Am. Chem. Soc. 141, 20470–20479 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.