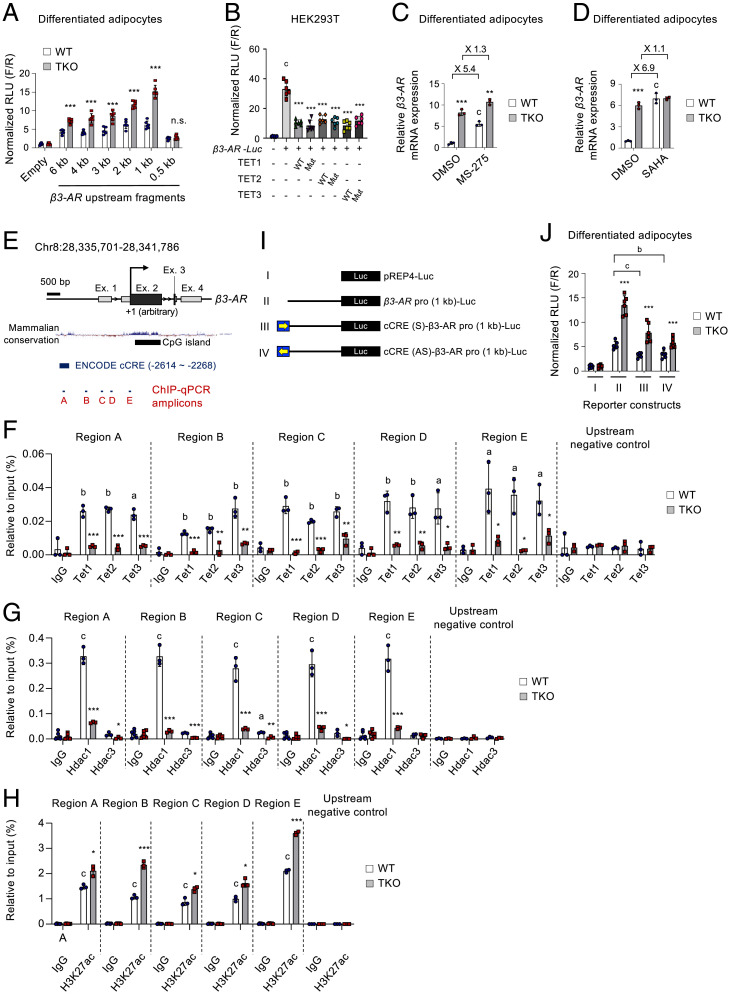
Fig. 7.
TET proteins cooperate with HDACs to directly suppress β3-AR transcription. (A) pREP4-based luciferase reporter activities driven by serially truncated β3-AR promoters in WT and Tet TKO adipocytes. Results were normalized to Renilla luciferase activity. n = 6. (B) pREP4-based luciferase reporter activities driven by the β3-AR (1 kb) promoter in HEK293T cells transfected with WT or mutant TET1, TET2, and TET3 plasmids. n = 6. (C and D) mRNA expression of β3-AR relative to Gapdh in WT and Tet TKO adipocytes stimulated with or without MS-275 (10 μM, 24 h) (C) or a pan-HDAC inhibitor, SAHA (20 μM, 6 h) (D). n = 3. (E) The genomic configuration of the mouse β3-AR gene locus. Locations of ChIP-qPCR amplicons, mammalian conservation, CpG islands, and Encyclopedia of DNA Elements (ENCODE) cis-CRE (cCRE) are shown. (F−H) ChIP-qPCR assays for Tet1, Tet2, and Tet3 (F); HDAC1 and HDAC3 (G); and acetylated H3K27 (H3K27ac) (H) for regions A−E shown in E. Enrichment at the nonbinding β3-AR upstream region was used as a negative control. n = 3. (I) Schematic representation of the β3-AR (1 kb) promoter-driven luciferase constructs. Yellow arrows indicate the direction of the incorporated ENCODE cCRE element shown in E. (J) pREP4-based luciferase reporter assay for the β3-AR promoters shown in (I) in WT and Tet TKO adipocytes. n = 6. All data are presented as the mean ± SD. The P values were determined by unpaired Student’s t test. *P < 0.05, **P < 0.005, ***P < 0.0005 versus WT; aP < 0.05, bP < 0.005, cP < 0.0005 versus reporter only (B), dimethyl sulfoxide (DMSO) (C), immunoglobulin G (IgG) (F–H), and reporter II in WT (J). Ex, exon; Mut, catalytically inactive mutant; n.s., not significant; RLU, relative light units; F/R, firefly/renilla.