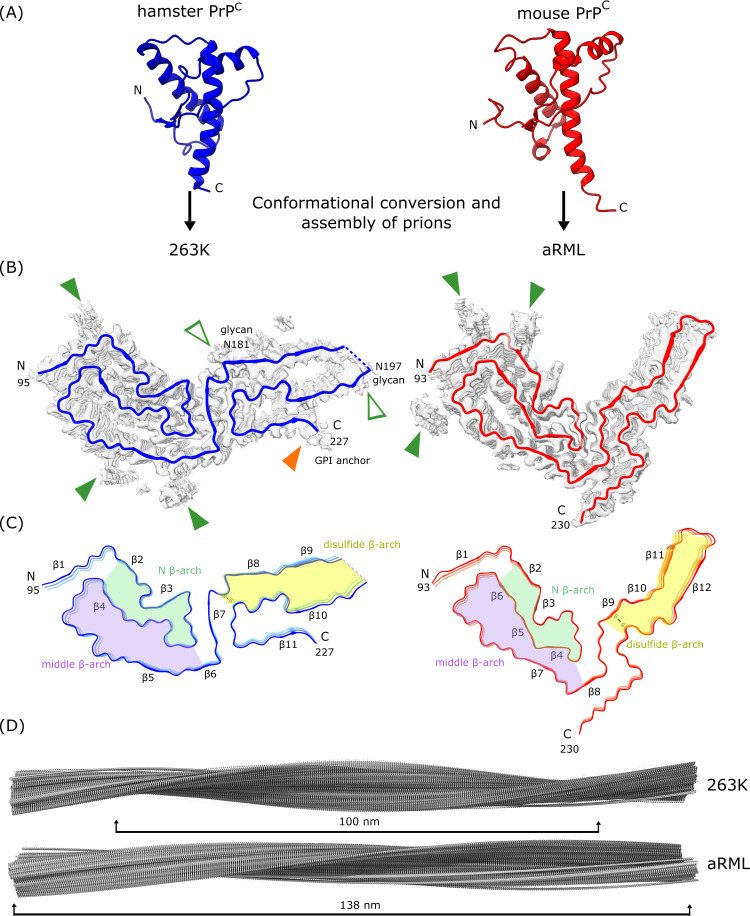
Fig 1. Two prion strains are amyloid fibrils with distinct templating surfaces on the ends where incoming monomers are refolded.
(A) PrPC molecules (disordered N-terminal residues not shown) undergo conversion and assembly into the prion forms shown here for hamster 263K (blue, PDB: 7LNA, [6,7]) and mouse aRML (red, PDB: 7TD6, [8,9]) prions. PDB accession codes 1B10 and 1XYZ were used for hamster and mouse PrPC, respectively. (B) Polypeptide backbones of the fibril cross-sections (colored) as they fit within the cryo-EM density maps (gray). Open arrowheads: densities outside the polypeptide backbone that coincide with attachment sites of glycans (green) or glycolipid anchors (orange). Closed green arrowheads: densities of unknown origin outside the amyloid cores, often adjacent to cationic residues. (C) Similar shared β-arch motifs/topologies distinguished by color. Variations within these topologies and other features, including distinct orientations of the C-terminal tails, distinguish these strains. (D) Extended lateral views of fibril density maps, with brackets indicating the relative cross-over distances. cryo-EM, cryogenic electron microscopy; PrP, prion protein.