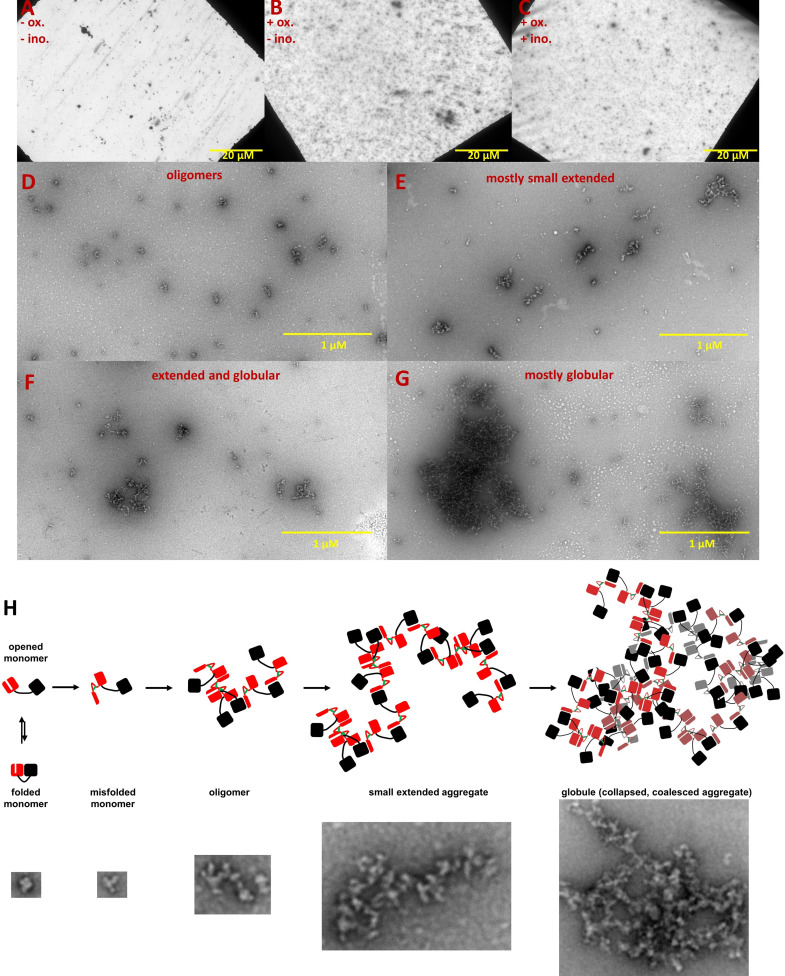
Figure 3. Negative-stain TEM images of HγD W42Q aggregates reveal short extended and large globular structures.
Aggregation of 40 μM protein in pH 6.7 PIPES buffer with 150 mM NaCl and 1 mM EDTA was triggered by addition of oxidant (0.5 mM GSSG) and incubation at 37 °C for 4 hr. End-point samples were deposited on carbon-coated copper grids and stained with uranyl acetate. The top row shows survey images of whole grids for (A) control sample incubated in the absence of GSSG; (B) turbid sample in the presence of GSSG; and (C) turbid sample in the presence of GSSG and 100 mM myo-inositol. Panels D-G show the variety of aggregate morphologies observed in representative magnified images from these grids, with (D) showing mostly small oligomers (not counted as aggregates in Figure 4); (E) showing mostly small extended aggregates; (F) showing extended aggregates in the process of collapsing and coalescing; and (G) showing highly coalesced globular aggregates; (H) Graphical model of HγD aggregation and its suppression by myo-inositol: aggregation begins with oxidative misfolding of a mutant or damaged protein, as we have previously found (Serebryany et al., 2016b), followed by assembly of short extended chains via domain swap-like interactions (Serebryany et al., 2016b; Serebryany et al., 2018), and finally coalescence to globular particles. Examples of TEM evidence of each type of structure in this study are shown in the bottom row (not all to scale).