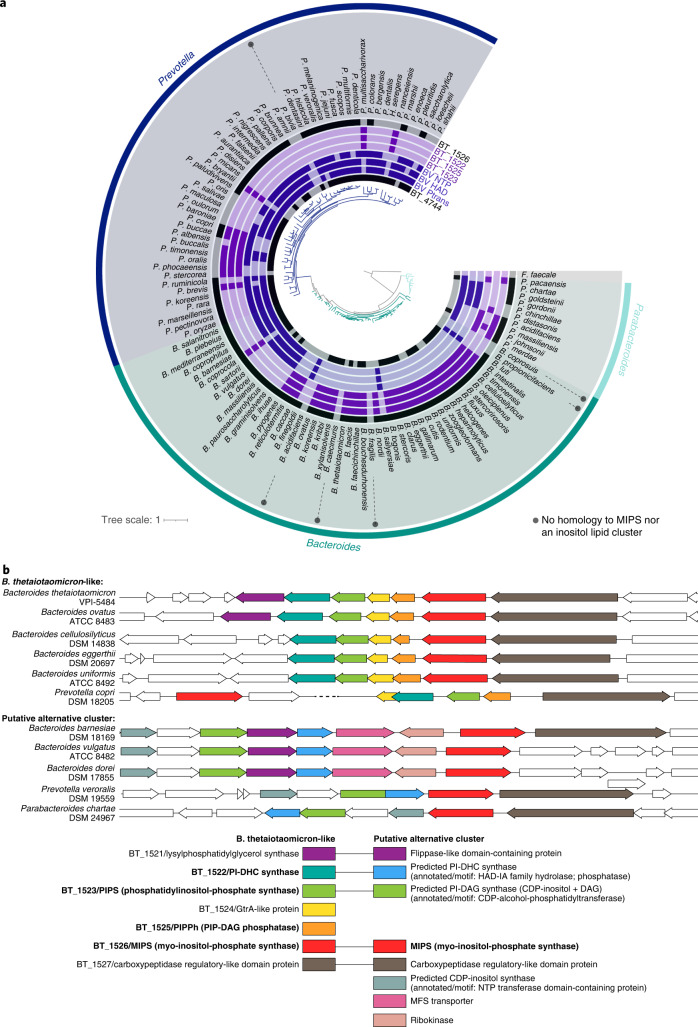
Fig. 6. The capacity for inositol lipid synthesis is widespread within the Bacteroidetes.
a, Maximum-likelihood-based phylogeny of representative Bacteroides, Prevotella and Parabacteroides species produced from 71 conserved single-copy genes present in all genomes (identified and concatenated using Anvi’o) and generated by RAxML (best tree; substitution model PROTCAT, matrix name DAYHOFF, Hill-climbing algorithm, bootstrap 50); Flavobacterium faecale is included as an outgroup. The rings surrounding the tree indicate species with genes that have NCBI BlastP homology to the BT inositol lipid cluster (in light purple; BT_1522, BT_1523, BT_1525, BT_1526), the BT Minpp (BT_4744), or representative proteins from the Bacteroides vulgatus putative alternative inositol lipid cluster (in dark purple; phosphatidyltransferase: BVU_RS13105 ‘BV Ptrans’; HAD hydrolase: BVU_RS13115 ‘BV HAD’; NTP transferase: BVU_RS13095 ‘BV NTP’). Homology at an e-value below 1 × 10−8 is indicated by dark coloration in the inner circles. b, Genomic regions surrounding the BT_1526/MIPS homologue in representative Bacteroidetes, compiled using the PATRIC 3.6.9 Compare Region Viewer. Protein homology (determined using NCBI BlastP) to proteins in the BT-like inositol lipid metabolism cluster (left in key) or the Bacteroides vulgatus-like putative alternative inositol metabolism cluster (right in key) is indicated by colour. The functions of enzymes in bold were characterized in this study; sequences with predicted redundant functions between both clusters are linked in the key.