Extended Data Fig. 1. XPR1 dependency is observed selectively in SLC34A2-high cancer cell lines.
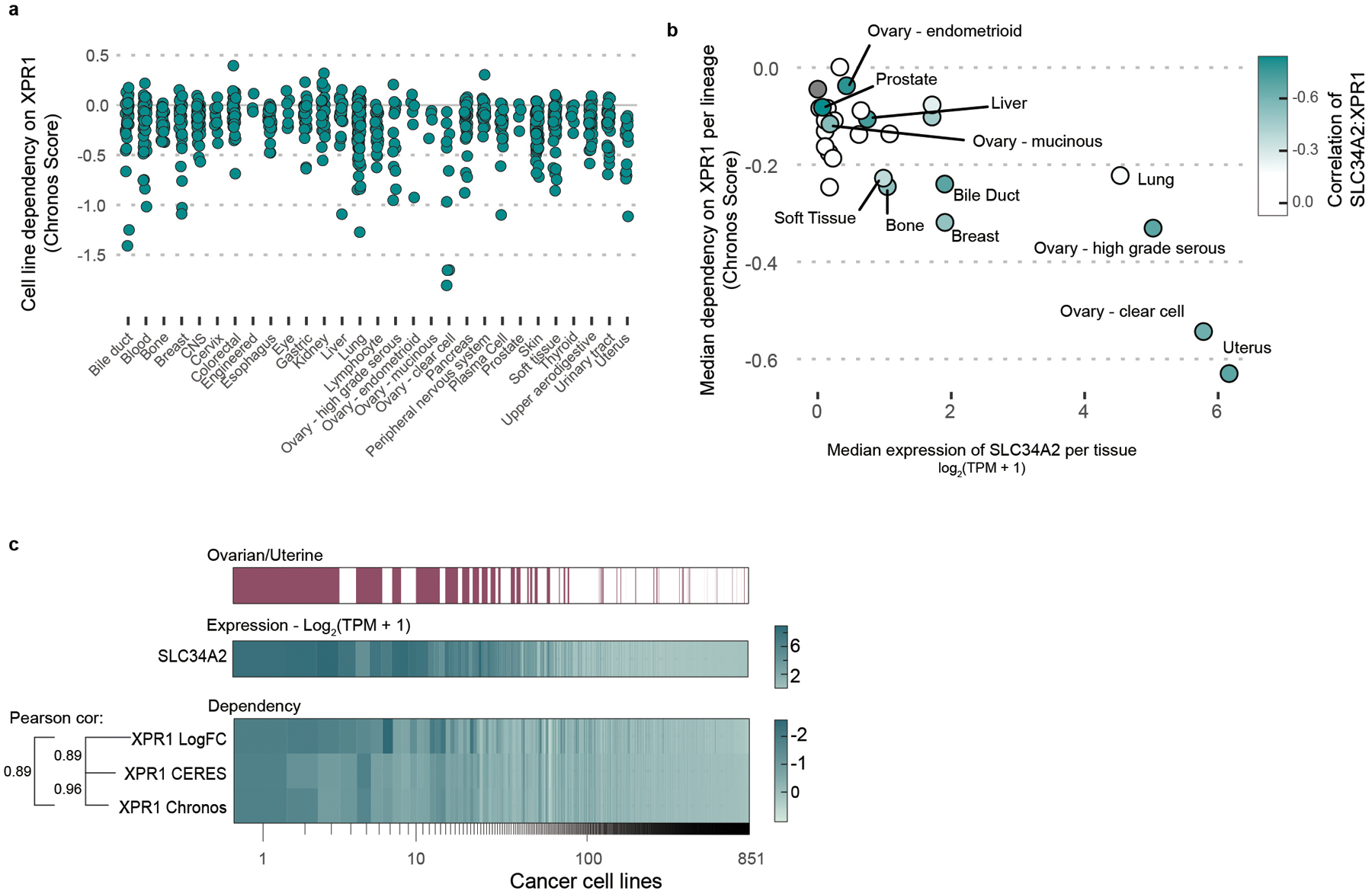
a) For every cell line profiled in the Cancer Dependency Map dataset (N=851 cancer cell lines), the degree of XPR1 essentiality is plotted on the Y-axis. The Chronos score is a scaled value of gene essentiality, where 0 is the effect of CRISPR/Cas9 genome editing and −1 is the effect of inactivation of pan-essential genes. Note that the ovarian lineage is separated into cancer subtypes.
b) For every tissue type, the 10 highest SLC34A2 expressing cell lines were analyzed for their median expression of SLC34A2 (X-axis) and dependency on XPR1 (Y-axis). Note that some lineages may have less than 10 cell lines. Color encodes the correlation of SLC34A2 expression and XPR1 dependency across all cell lines within that lineage.
c) Comparison of analytical methods for CRISPR/Cas9 genome-scale loss of function screens.
