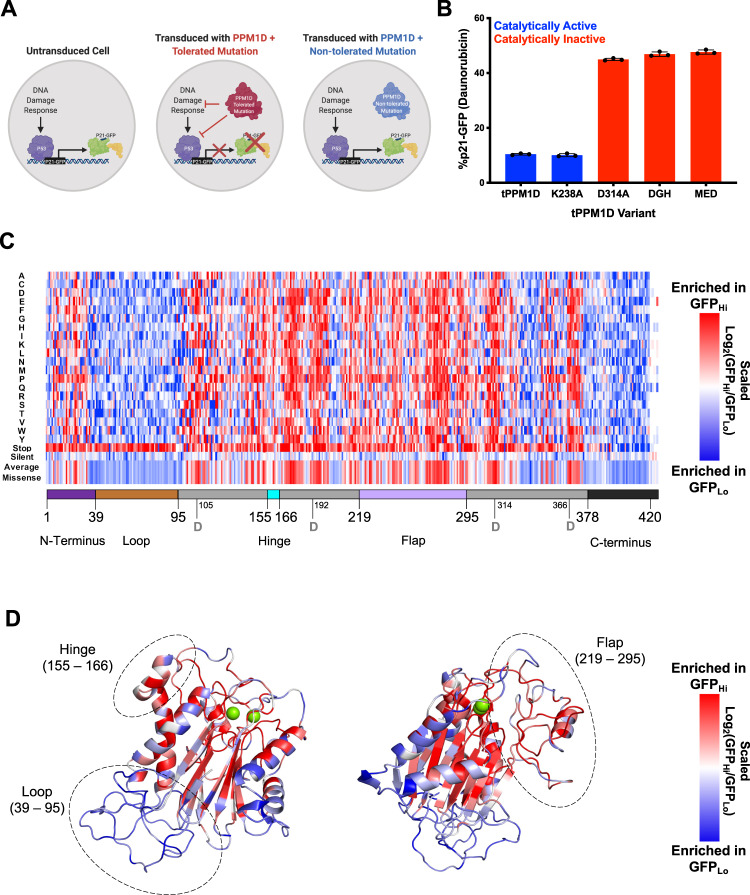
Fig. 2. Deep mutational scanning identifies residues important for PPM1D function.
A Schematic of engineered K562 cells with wild-type TP53 and GFP knocked into the C-terminus of the CDKN1A (p21) locus. In the presence of a DNA damaging agent, the DNA damage response (DDR) pathway induces p53 activation which transactivates the p21 locus, leading to expression of p21-GFP. After treatment with a DNA damaging agent such as daunorubicin, cells expressing a truncated PPM1D with a mutation that does not impair the function of the protein (red) suppresses the DDR and GFP expression whereas cells expressing truncated PPM1D that does impair protein function (blue) does not suppress the DDR or GFP expression. B Effect of mutations on PPM1D-mediated suppression of the DDR. Cells expressing truncated PPM1D (tPPM1D) or a mutation that does not affect protein function (tPPM1D-K238A) have lower p21-GFP levels at baseline and in the presence of daunorubicin. In contrast, cells expressing truncated PPM1D with mutations that impair catalytic function (tPPM1D-D314A, tPPM1D-DGH(105-107)→AAA, or tPPM1D-MED(21–23)→AAA)17 have higher levels of p21-GFP at baseline and in the presence of daunorubicin. N = 3 biologically independent samples. Data are presented as mean values ± SEM. Source data are provided as a Source Data file. C Representation of deep mutagenesis screen. Each column represents the amino acid position and each row represents that amino acid change in the variant including stop codons, silent mutations, or the average of all the missense mutations. For each variant the log2(GFPHi/GFPLo) was calculated and scaled with red representing a greater ratio (impaired activity), and blue representing a lesser ratio (retained activity). See Supplemental Table 4 for source data. D Overlap of heatmap missense mutations on theoretical model. The average effect of the missense variants at each amino acid position are overlaid onto the theoretical model of truncated PPM1D.