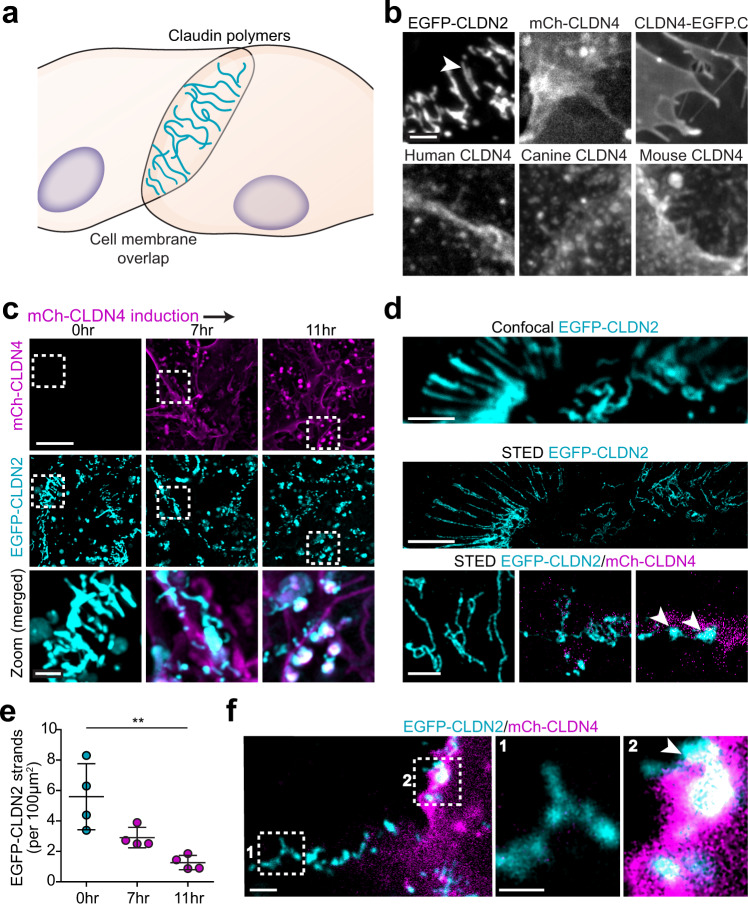
Fig. 5. Claudin-4 fails to assemble into polymeric strands but disrupts those formed by claudin-2.
a Claudin proteins expressed in nonepithelial cells form polymers similar to those within epithelial tight junction strands. b EGFP-claudin-2 expressed in U2OS human osteosarcoma cells forms strands (arrowhead). In contrast, mCherry-claudin-4 (human), claudin-4-EGFP (CLDN4-EGFP.C), untagged human claudin-4, canine claudin-4, and mouse claudin-4 all fail to form strands despite being trafficked to plasma membranes. Maximum projection images are representative of 3 independent experiments. Scale: 2 μm. c Live imaging at intervals after induction of mCherry-claudin-4 (magenta) expression shows progressive disruption of EGFP-claudin-2 (cyan) strands. Maximum projection images representative of 5 independent experiments. Scale: 10 μm; 2 μm. d Confocal and corresponding STED image of EGFP-claudin-2 polymers (cyan) without claudin-4 expression. STED shows collapse of EGFP-claudin-2 (cyan) polymers into dense aggregates (arrowhead) following mCherry-claudin-4 (magenta) expression (bottom). Representative of at least 7 independent experiments. Scale: 1 µm. e Quantitative morphometry shows progressive loss of EGFP-claudin-2 strands as mCherry-claudin-4 expression increases. n = 4 (each point is the mean of 12–16 cells in 1 sample), representative of 6 independent experiments. 1-way ANOVA. **P = 0.0035. f EGFP-claudin-2 strands (box 1) are replaced by amorphous aggregates at sites of asymmetric mCherry-claudin-4 expression (box 2), suggesting that cis interactions are sufficient for claudin-4-mediated disruption of claudin-2 strands. Maximum projection images representative of 8 independent experiments. Scale: 1 µm. Data are presented as mean ± SD and included in the Source Data file.