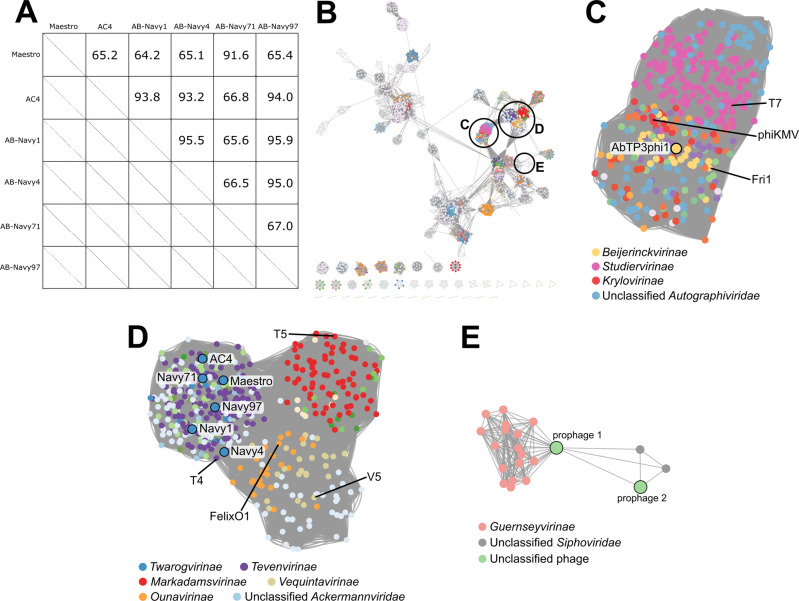
Fig. 1. DNA and protein-based relationships of the seven treatment phages.
A DNA sequence relatedness of six T4-like myophages, showing pairwise percent DNA sequence identities as determined by ProgressiveMauve (upper section) and DNA dotplots visually representing DNA sequence alignments between phages (lower section). B Protein sequence-based relationships of 2834 Caudoviricetes phages representing all species in the ICTV taxonomy, plus the seven treatment phages and two prophages identified in strains TP1, TP2, and TP3. Unclustered singletons (17 in total) are removed from the visualization. Distinct colors are assigned at the Subfamily level; if no Subfamily was assigned, color is assigned at the Family level. Circled clusters are enlarged in C–E as labeled. C Enlarged cluster representing the Autographiviridae. Nodes are colored based on their Subfamily membership, with the legend identifying prominent clades; the node representing phage AbTP3phi1 is outlined in black, and nodes representing clade-founding phages T7, phiKMV and Fri1 are labeled. D Enlarged cluster containing the T4-like subfamilies, including the Twarogvirinae; this large cluster is also linked to the T5-like Markadamsvirinae and clusters of diverse myophages including the V5-like Vequintaviridae and FelixO1-like Ounavirinae. Nodes are colored based on their Subfamily membership with the legend identifying prominent clades. Nodes representing the six treatment myophages are outlined in black, and nodes representing clade-founding phages T4, T5, FelixO1 and V5 are labeled. E Enlarged cluster containing the two prophage elements identified in strains TP1, TP2 and TP3. These prophages are not closely related to other classified phages, with the 52 kb prophage 1 distantly linked to the Guernseyvirinae, and the 42 kb prophage 2 related to two other unclassified siphophages.