Figure 1.
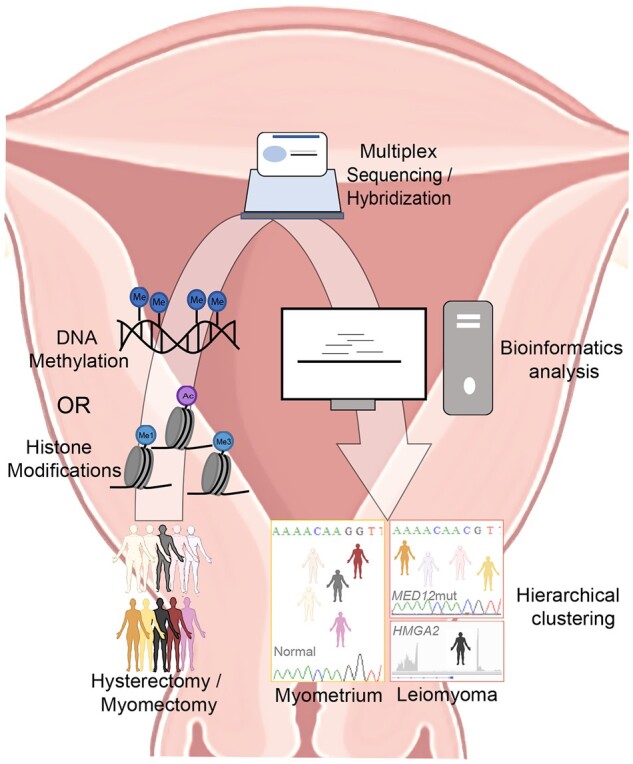
Schematic illustration of workflow employed to identify histone modification/DNA methylation patterns that discriminate uterine leiomyoma from normal myometrium. Tissue samples from patients undergoing hysterectomy or myomectomy were frozen until use. Following appropriate processing, samples were prepared either for ChIP-sequencing (histone modifications, Leistico et al., 2021) or hybridization (DNA methylation, George et al., 2019). Data were analyzed using various bioinformatic tools and unsupervised clustering was performed. Epigenomic alterations could cluster myometrium separately from uterine leiomyoma [LM] and, furthermore, LM into subtypes (MED12mut indicates mediator complex subunit 12 mutated LM subtype; HMGA2, overexpressed high mobility group AT-hook 2 LM subtype).
