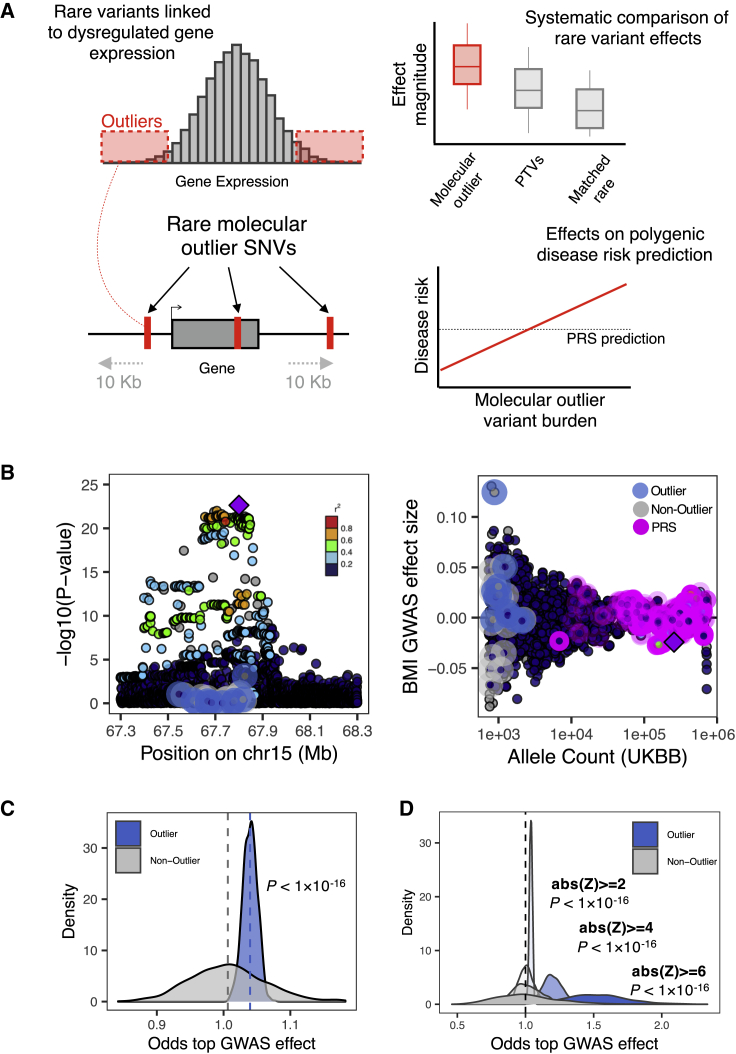
Figure 1.
Phenotypic effects of rare outlier-associated variants across genes
(A) Rare SNVs were identified in gene expression outlier individuals across 49 GTEx v8 tissues. The phenotypic effects of these variants were systematically compared with protein-truncating variants and matched rare non-outlier variants and jointly modeled with PRS estimates.
(B) Example gene locus (MAP2K5) containing a common variant genome-wide significant hit for BMI illustrates the large phenotypic effect of an outlier-associated variant: (left) showing distribution of −log10(p values) for UKB BMI GWAS for all outlier (blue halo) and non-outlier (gray halo) variants linked to the gene; (right) associated effect sizes, stratified by UKB allele count and highlighting variants included in a PRS for BMI (pink halo). Points colored by LD (1000 Genomes phase 3, European samples) relative to lead variant in gene locus (purple diamond).
(C) Distribution of odds estimates from permutation testing to assess how often randomly drawn outlier-associated variants had larger BMI GWAS effect sizes than matched non-outlier variants across genes (blue shading). This process was repeated for randomly selected non-outlier variants only (gray). p values obtained with a Wilcoxon test.
(D) Distribution of odds from permutation testing (permutation testing method as detailed in B), across more-stringent outlier Z scores. p values obtained with a Wilcoxon test.