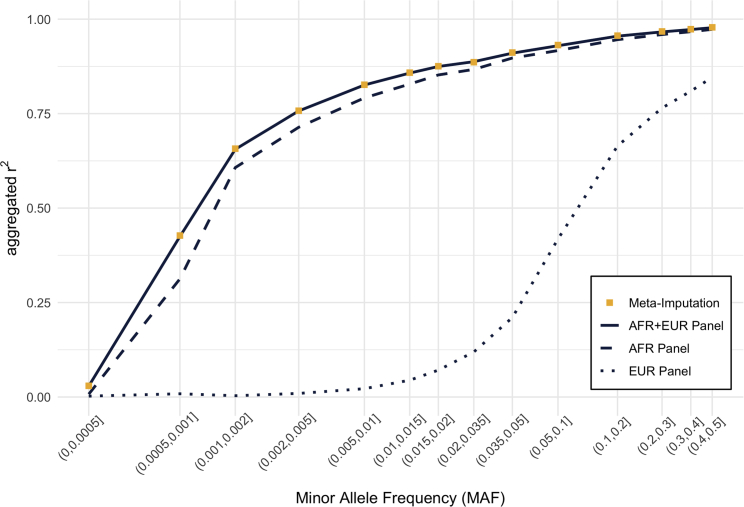
Figure 2.
Comparison of imputation accuracy in African American samples
Imputation accuracy for the pseudo-GWAS ASW dataset was compared among (1) meta-imputation, (2) imputation using the combined AFR + EUR panel including both African and European ancestry genomes, (3) imputation using the homogeneous African (AFR) panel, and (4) imputation using the homogeneous European (EUR) panel. Variants were grouped according to minor allele frequency, which was estimated from the genotype data of 2,504 samples in the 1000 Genomes Project. Aggregated r2 values were calculated for each variant group.