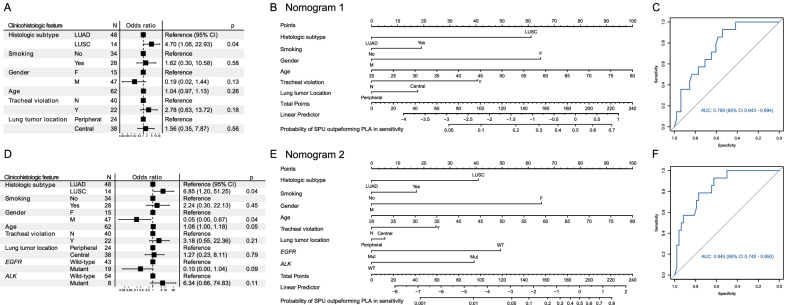
Fig. 5.
Nomograms for predicting the probability of achieving more sensitive genome profiling with sputum supernatant than with plasma based on clinicohistologic features with A–C unknown or D–F known mutation status of EGFR or ALK. A, D Forest plots of multivariate analysis for clinicohistologic characteristics associated with the probability of greater sensitivity with SPU than with PLA. B, E Nomograms constructed with features shown in (A and D). To use the nomograms, vertical lines connecting the values of each variable with the point score are first drawn at the top of the diagram. The scores for each variable are then summed to give a total points score, which is plotted along the “total points” line near the bottom. A vertical line drawn downward through scale at the bottom allows calculation of the probabilities of more sensitive genome profiling with sputum supernatant than with plasma. A worked example of (B) can be found in Additional file 1: Fig. S2. C, F Receiver operating characteristic analysis for validating the discrimination power of the nomograms. AUC area under curve, CI confidence interval, SPU sputum supernatant, PLA plasma