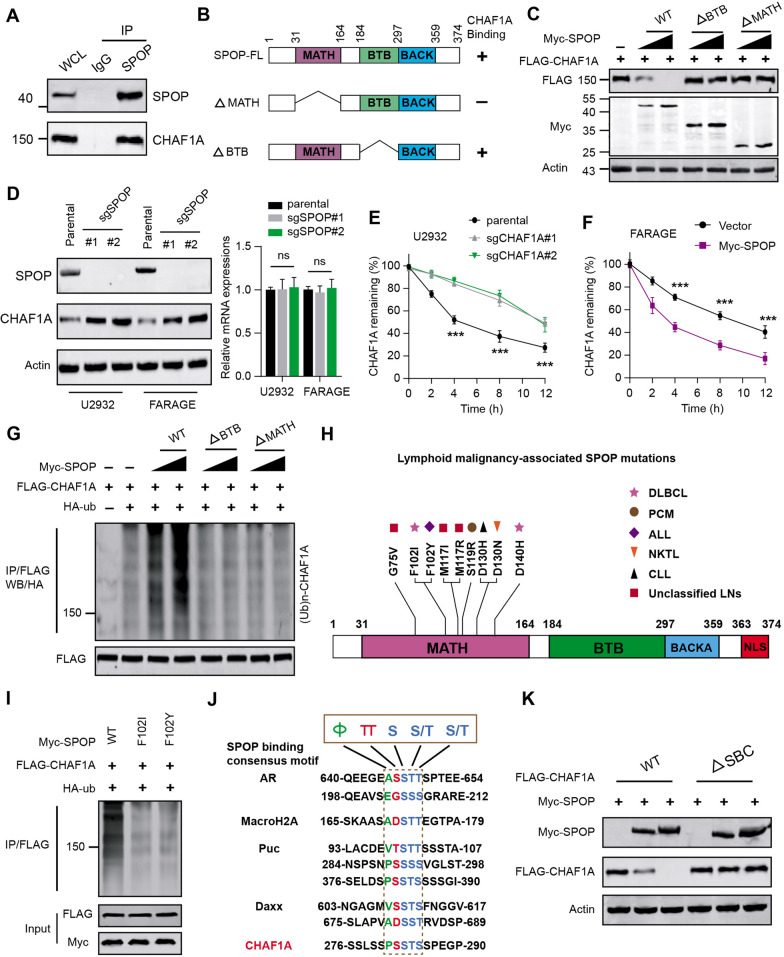
Fig. 3.
Aberrant SPOP-CHAF1A ubiquitination axis contributes to accumulated CHAF1A expressions in DLBCL. A Co-Immunoprecipitation (IP) analysis based on U2932 cells with IgG or anti-SPOP antibody indicated the endogeous interactions between SPOP and CHAF1A, as shown by western blotting assay. B The schematic representation of SPOP-WT and deletion mutants. Binding capacity of SPOP to CHAF1A is indicated with the representative symbol. C Western blotting assay showed the altered CHAF1A proteins in whole cell lysate (WCLs) from U2932 cells which were co-transfected with plasmids of FLAG-CHAF1A and WT SPOP or mutants. D Western blotting analysis showed the elevated CHAF1A proteins in CHAF1A KD cells (U2932, FARAGE) relative to control cells. Besides, the corresponding mRNA levels of CHAF1A in indicated groups were quantified and shown on the right, and no altereations were observed. E Quantification of CHAF1A proteins in WCLs of U2932 cells (parental, sgSPOP) for 12 h and then treated with 50 μg/ml cycloheximide (CHX) and harvested at different time points. At each time point, the intensity of CHAF1A was normalized to the intensity of actin and then to the data at 0 h. F Quantification of CHAF1A proteins in WCLs of FARAGE cells (EV, myc-SPOP) for 12 h and then treated with 50 μg/ml cycloheximide (CHX) and harvested at different time points. G After the 293 T cells were transfected with the indicated plasmids and treated with 20 μM MG132 for 8 h, the western blotting assay showed the in vivo CHAF1A ubiquitination levels by WT SPOP or mutants. H Illustration of tumor-associated mutations across SPOP gene in lymphoid malignancies. DLBCL: diffuse large B Cell lymphoma; PCM: plasma cell myeloma; ALL: acute lymphoid leukemia; NKTL: NK-T cell lymphoma; CLL: chronic lymphocytic leukemia; unclassified LNs unclassified lymphoid malignancies. I Western blotting assay showed the differential in vivo ubiquitination levels of CHAF1A in 293 T cells transfected with plasmids of FLAG-CHAF1A and Myc-SPOP or relevant mutations. J Comparison of putative SPOP binding sites in CHAF1A with the SPOP-binding consensus motif defined in the known SPOP substrates, where amino acid sequence alignment of the SBC motif (Φ-π-S-S/T-S/T; Φ: nonpolar residues, π: polar residues) was identified. K Western blotting assay showed the altered protein levels of WT-CHAF1A and mutants with deleted SBC motif when U2932 cells were transfected with Myc-SPOP, respectively. Experiments were performed in triplicate. *p < 0.05, **p < 0.01, ***p < 0.001