Fig. 1 |. Alignment of insulin sequences.
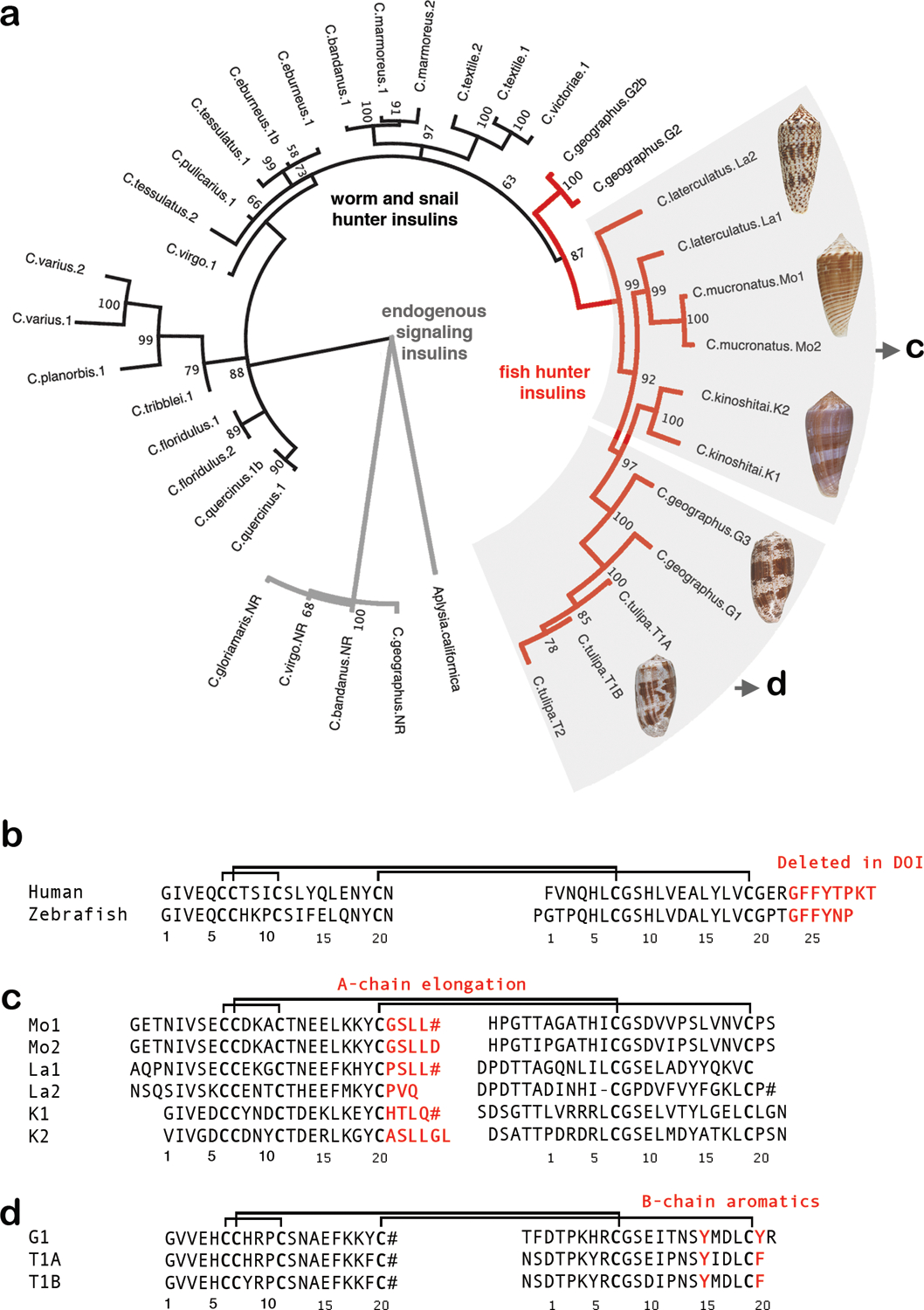
a, Molecular phylogenetics closely groups the newly identified venom insulin sequences with venom insulins previously identified from other fish hunters (red branches in tree). Tree branches of venom insulins from snail and worm-hunters are shown in blue, and those of endogenous signaling insulins are black. b, Sequence alignment of human and zebrafish insulin c, Alignment of venom insulins sequenced here from C. mucronatus (Mo1 and Mo2) and C. laterculatus (La1 and La2) and previously from C. kinoshitai10 (K1 and K2). d, Alignment of venom insulins from C. geographus (G1) and C. tulipa (T1A and T1B)8. B-chain residues deleted in DOI and important for receptor-binding and dimerization (b) or residues unique to venoms and predicted to bind insulin receptor (c and d) are in red. Cysteines are in bold. Disulfide connectivity is shown as black lines. # indicates C-terminal amides.
