Fig. 3 |. The symmetric Vh-Ins-HSLQ-receptor structure.
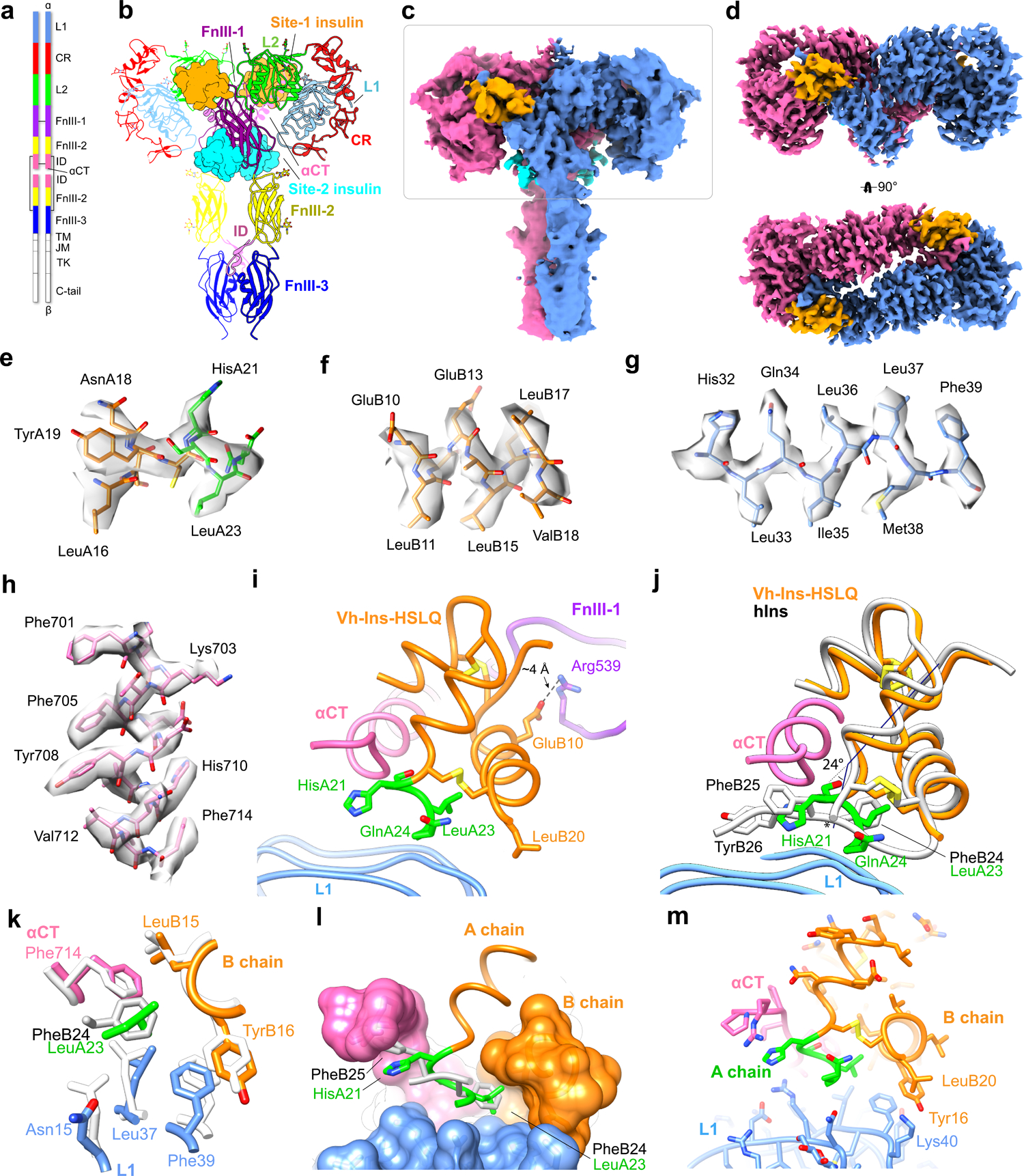
a, Schematic of the insulin receptor domains and disulfide connectivity. b, Structure of the insulin receptor ectodomain with four Vh-Ins-HSLQ molecules bound. Insulins are depicted in surface representation (orange, site 1; cyan, site 2). c, Consensus refinement density of the Vh-Ins-HSLQ-receptor complex (4.1 Å). Box indicates the region selected for focused refinement. Blue and pink, receptor protomers. Vh-Ins-HSLQ orange (site 1) and cyan (site 2). d, Focused refinement map. Site-2 insulins do not contribute significant signal relative to noise at the filter frequency used for the final reconstruction (3.4 Å). e-h, representative density and model for Vh-Ins A chain, Vh-Ins B chain, L1, and αCT residues, respectively. i, The extended A-chain residues (green) are in close proximity to the receptor αCT and L1 domains. LeuB20 contacts L1 and GluB10 interacts with FnIII-1. j, Comparison of site-1 Vh-Ins-HSLQ (orange) and human insulin (white, PDB 6PXW)16 aligned by superposition of L1 and αCT residues. Vn-Ins-HSLQ extended A-chain residues (green) contact the same receptor surface engaged by human insulin PheB24 and PheB25. The helix formed by Vh-Ins-HSLQ residues A13-A23 is kinked 24 ° away from αCT at HisA21 and slightly unwound. The human insulin A-chain C terminus is indicated with an asterisk. k, Vh-Ins-HSLQ LeuA23 binds a hydrophobic pocket in a similar fashion to human insulin PheB24. l, Surface representation of the pocket formed by αCT and L1 and binding by LeuA23 and PheB24. m, Vh-Ins-HSLQ LeuB20 packs against TyrB16 and approaches receptor Lys40.
