FIGURE 2.
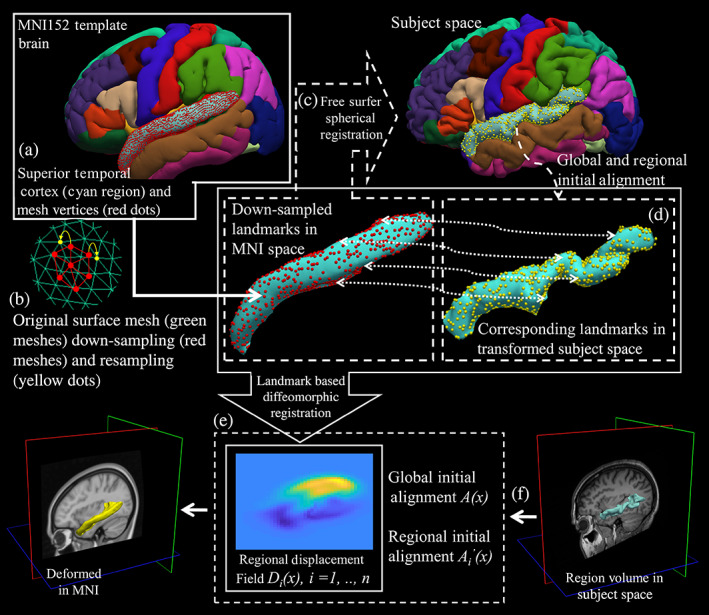
Illustrate our method for automatic landmark extraction and matching for landmark‐based regional nonlinear registration with example on superior temporal cortex (STC) region. In step (a), for STC region (cyan color), vertices of the GW/WM and GM/CSF boundaries triangular meshes are extracted as landmarks (GM/CSF surface vertices shown as red dots, GW/WM surface vertices are not showing in the figure); In step (b), landmarks of STC region are down‐sampled by down‐sampling the original dense surface mesh (green meshes) to a sparser surface mesh (red meshes) and sampled back to the original vertices (yellow dots) to keep consistency; In step (c), correspondence of STC regional landmarks between the MNI template and the subject is established through spherical registration (corresponding landmarks in subject space shown as yellow dots); In step (d), corresponding regional landmarks are initially aligned with linear transformations in 3D Euclidean space; In step (e), a diffeomorphic nonlinear landmark‐based registration is used to generate regional warping field for STC region. The step (f) is only showing that STC regional warping field can be used to warp the subject's regional volume onto MNI template space. CSF, cerebrospinal fluid; GW, gray matter; WM: white matter
