Abstract
Microscopy and genomic approaches provide detailed descriptions of the three-dimensional folding of chromosomes and nuclear organization. The fundamental question is how activity of molecules at the nanometer scale can lead to complex and orchestrated spatial organization at the scale of chromosomes and the whole nucleus. At least three key mechanisms can bridge across scales: (1) tethering of specific loci to nuclear landmarks leads to massive reorganization of the nucleus; (2) spatial compartmentalization of chromatin, which is driven by molecular affinities, results in spatial isolation of active and inactive chromatin; and (3) loop extrusion activity of SMC (structural maintenance of chromosome) complexes can explain many features of interphase chromatin folding and underlies key phenomena during mitosis. Interestingly, many features of chromosome organization ultimately result from collective action and the interplay between these mechanisms, and are further modulated by transcription and topological constraints. Finally, we highlight some outstanding questions that are critical for our understanding of nuclear organization and function. We believe many of these questions can be answered in the coming years.
THREE MECHANISMS OF CHROMOSOME FOLDING AND THEIR INTERPLAY IN NUCLEAR ORGANIZATION
The organization of the cell nucleus is directly related to the folding of chromosomes and their associations with subnuclear structures such as the nuclear lamina. Microscopy and genomic approaches now allow detailed descriptions of the three-dimensional (3D) organization of chromosomes, the presence of nuclear bodies such as the nucleolus, speckles, and Cajal bodies around specific loci, and the formation of subnuclear compartments enriched in sets of loci and specific trans factors. Principles of genome organization, the dynamics in this organization during the cell cycle (Dileep et al. 2015; Gibcus et al. 2018; Abramo et al. 2019; Zhang et al. 2019; Kang et al. 2020) and development (Hug et al. 2017; Wike et al. 2021), and variation in folding between single cells (Nagano et al. 2013; Ramani et al. 2017) are now being described at increasing resolution.
In recent years, at least three mechanisms that contribute to chromosome folding and nuclear organization have come into focus, allowing going beyond descriptive studies (Fig. 1). First, loci can be tethered to specific nuclear features such as the nuclear periphery (Guelen et al. 2008). For instance, genetic perturbation experiments have identified specific factors (e.g., lamin B receptor and lamin A/C) required for the tethering of heterochromatic loci to the nuclear lamina (Solovei et al. 2013). Second, the nucleus is compartmentalized so that active and inactive chromatin domains are spatially segregated (Simonis et al. 2006; Lieberman-Aiden et al. 2009). Interestingly, the spatial segregation of active and inactive chromatin is not dependent on tethering of heterochromatin and occurs also in inverted nuclei in which such tethering is absent and heterochromatin is clustered in the center of the nucleus (Solovei et al. 2009; Falk et al. 2019). The spatial separation of active and inactive chromatin (compartmentalization) and the formation of subnuclear bodies both can be understood as the result of phase separation (Falk et al. 2019; Hildebrand and Dekker 2020). This biophysical process can drive clustering of specific types of chromatin and aggregation of sets of proteins through weak multivalent interactions. Theoretical considerations that such a process could explain aspects of nuclear organization are now leading to testable predictions of how experimental perturbations would alter compartmentalization. Third, chromatin is folded into dynamically growing and disappearing loops. Theoretical considerations predicted that specific molecular machines form chromatin loops through a process of loop extrusion (Fudenberg et al. 2016, 2017). This mechanism is increasingly understood in molecular detail. Loop extrusion activity has been directly observed for the SMC (structural maintenance of chromosome) complexes cohesin and condensin that can extrude chromatin loops in an ATP-dependent manner (Ganji et al. 2018; Davidson et al. 2019; Kim et al. 2019; Golfier et al. 2020; Kong et al. 2020). Loop extrusion can explain many features of chromatin folding in interphase and in mitosis.
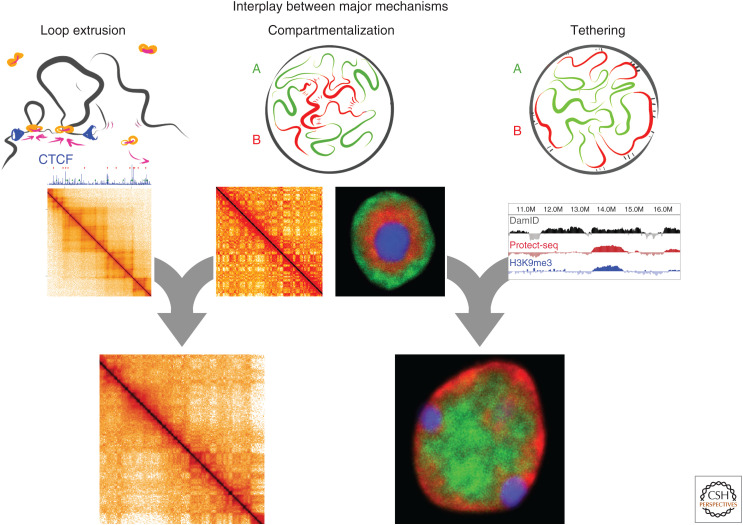
Figure 1.
Summary of how the interplay between major mechanisms of chromosome folding results in nuclear organization. Loop extrusion (left column) occluded by CTCF results in topologically associating domains (TADs), stripes, and dots, but also influences compartmentalization of euchromatin and heterochromatin. (Middle column) In the absence of extrusion (owing to cohesin depletion), compartmentalization gets stronger and finer, and better follows patterns of histone modifications. Compartmentalization observed in wild-type cells (bottom row) is a result of the interplay between such modification-dependent compartmentalization and loop extrusion. In the absence of tethering, attractions between heterochromatic (red) regions result in a phase-separated but inverted nucleus (middle column) in which constitutive heterochromatin (blue) is located in the center of the nucleus, surrounded by the facultative heterochromatin (B compartment, red), with euchromatin (A compartment) at the nuclear periphery. Lamina tethering, in turn, leads to peripheral location of heterochromatin as evident from DamID and Protect-seq (Spracklin and Pradhan 2020). The interplay of attraction between regions of heterochromatin and its tethering to the nuclear lamina results in conventional nuclear organization. (Hi-C data from Falk et al. 2019; microscopy images courtesy of Irina Solovei.)
Ultimately, the folded state of the genome, and the organization of the nucleus in general, is the result of interplay between different biophysical and molecular mechanisms that fold chromosomes. For instance, tethering and phase separation together result in the spatial organization of heterochromatin and euchromatin in the nucleus (Falk et al. 2019). Another example discussed below is the interference of loop extrusion with compartmentalization (Schwarzer et al. 2017; Nuebler et al. 2018). Moreover, different loop extruding complexes can interfere with each other (e.g., condensin and cohesin), and the relative contributions of the two condensin complexes determine whether mitotic chromosomes are long and thin, or short and wide (Shintomi and Hirano 2011).
In this review, we outline the different mechanisms that fold chromosomes, and focus on their interplay that results in the folding patterns observed in the cell. We also highlight outstanding questions. Given the enormous progress over the past years in development of both experimental methods and powerful predictive theoretical models, we believe many of these questions can be answered in the coming years.
COMPARTMENTALIZATION
One of the most prominent patterns in Hi-C data of higher eukaryotes is the checkerboard pattern that consists of ∼0.1–2 Mb size rectangular “checkers” visible in both intrachromosomal (cis) and interchromosomal (trans) sections of Hi-C maps (Lieberman-Aiden et al. 2009). Prominent during interphase, the checkerboard patterns rapidly (<15 min) disappear when cells go into mitosis (Gibcus et al. 2018), and gradually reemerge (∼2–4 h) after exit from mitosis (Abramo et al. 2019; Zhang et al. 2019). Although weak during early stages of embryonic development (Du et al. 2017; Ke et al. 2017), the checkerboard pattern appears as early as in mouse zygotes (Flyamer et al. 2017) and increases in intensity through early development. Interestingly, the pattern is present in the paternal, but not in the maternal zygotic pronuclei. Compartments are absent in yeast and in bacteria. Compartmentalization in archaea has been reported by some groups (Takemata et al. 2019), whereas others (Cockram et al. 2021) suggested that domains in archaea are of a different nature. Positions and intensity of the checkers vary between metazoan cell types suggesting their connection to transcriptional and/or chromatin states of the genome.
This checkerboard pattern reflects the presence of two (or more) types of genomic regions that alternate along the chromosomes and have an enrichment of interactions between regions of the same type and depletion of interactions between regions of different types. Such alternating regions are referred to as compartments, compartmental domains, or compartmental regions, and the pattern of preferential interactions between them is referred to as compartmentalization.
Genomic locations of compartmental regions can be directly inferred from Hi-C maps. One approach is to infer the “compartment genomic track” is by eigenvector decomposition (Lieberman-Aiden et al. 2009; Imakaev et al. 2012). Identified this way, compartments show a striking correlation with local transcriptional activity and specific histone marks. Regions of one type (compartment A) correspond to transcriptionally active and open regions of the genome, and the other type (compartment B) corresponds to silent, gene-poor, or repressed regions (Lieberman-Aiden et al. 2009). Regions of type A are rich in marks of active chromatin: H3K27ac, H3K4me1/3, H3K36me3, etc., whereas B regions are enriched in inactive marks (e.g.,H3K9me2/3). As such, A compartments closely match euchromatin and B matches heterochromatin. Note that repeat-rich parts of heterochromatin, typically pericentromeric and peri-telomeric, are not visible in Hi-C, and hence cannot be classified as B compartment. Compartmentalization is also highly correlated with features of genomic sequence that are known to be associated with hetero- and euchromatin (e.g., GC composition in warm-blooded vertebrates [high in A, lower in B], short interspersed nuclear elements [SINEs] associated with A compartment, and long interspersed nuclear elements [LINEs] with B compartment) (Solovei et al. 2016). Nevertheless, the transcriptional and epigenetic states of a locus determine its compartmental type; hence, A/B compartments can be different in different cell types.
Enrichment of contacts between compartmental regions of the same type as seen in Hi-C, reflect spatial segregation of euchromatin (A compartment) and heterochromatin (B compartment). In the vast majority of nuclei, euchromatin occupies the center of the nucleus, and heterochromatin is located at the nuclear periphery and surrounds nucleoli (Solovei et al. 2016; van Steensel and Belmont 2017).
What mechanisms can guide spatial segregation of A and B regions? Such spatial segregation is reminiscent of phase separation in polymers made of A- and B-type monomers with A-A and/or B-B attraction (Rubinstein and Colby 2003). Consistently, several studies showed that the spatial segregation of compartments and the checkerboard patterns observed in Hi-C can be reproduced in polymer simulations in which regions of the same type attract each other (some studies erroneously referred to such regions as topologically associating domains [TADs], not compartments) and in which B regions were additionally tethered to the lamina (Barbieri et al. 2012; Jerabek and Heermann 2012; Jost et al. 2014; Di Pierro et al. 2016; MacPherson et al. 2018). A recent study (Falk et al. 2019) allowed to disentangle contributions of these factors indicating that compartmentalization is mostly driven by strong attractions between B regions, with weaker attractions between A regions, and is independent from tethered to the lamina (see below).
Not all regions, however, fall under the simple A/B classification. Regions with an intermediate value of the compartment signal (the first eigenvector) seem to avoid interactions with either A or B, likely forming a separate compartment (referred as I compartment) (Schwarzer et al. 2017; Johnstone et al. 2020). Another interesting exception are Polycomb-repressed H3K27me3 regions that tend to interact with each other (Vieux-Rochas et al. 2015; Boyle et al. 2020), but that can show little enrichment in interactions with either A or B compartments, and are spatially located within the central (euchromatic/A compartment) part of the nuclear volume (Girelli et al. 2020). Another pattern that stands out from the A/B compartmentalization is the enrichment of interactions between exon-rich genes, likely mediated by interactions with splicing factories (Bonev et al. 2017; Kerpedjiev et al. 2018; Zhang et al. 2021). This observation implies that the process of transcription and splicing can contribute to at least some aspects of nuclear compartmentalization. Likely more exceptions from the A/B compartmentalization will be discovered, suggesting that more compartment types will be necessary to describe them.
Several questions about mechanisms and functional role of compartmentalization remain open:
What is the functional role of spatial segregation between different types of genomic regions? Is spatial compartmentalization necessary for their function: gene silencing in B and gene activation in A, important for the maintenance of histone marks, or necessary for some other genome maintenance functions (e.g., silencing of transposable elements)?
How do repressive compartments repress? Which factors are critical for this: high volume-density of heterochromatin, its peripheral location, or presence of specific histone marks, DNA modification, or recruitment of repressors?
What molecular players are key for mediating attractions between different types of chromatin? Can affinities be driven by direct interactions between methylated histones or do they require “bridging” by proteins (e.g., PRC and HP1 family) or RNAs?
How many types of compartmental regions, beyond eu-/heterochromatin are present in the cell? Can some compartment types emerge or become widespread only in specific cell types? What are their mutual interactions? What functional roles do multitudes of compartment types play?
Is compartmentalization a result of transcription and histone modifications or is it a driver for establishment of transcription and histone modification patterns? Are there feedback mechanisms between compartmentalization and processes on the DNA? Does compartmentalization help to “memorize” the transcriptional state of a cell or does it drive changes in cellular states?
What is the evolutionary origin of compartmental organization? Is it essential for establishment and maintenance of the many different cell types in multicellular organisms?
LOOP EXTRUSION
During interphase, Hi-C maps of higher eukaryotes show rich patterns of contact enrichments near the diagonal (i.e., between regions separated by less than a few megabases) (Hsieh et al. 2020; Krietenstein et al. 2020). When first characterized in 2012, such patterns were limited by the resolution of the data (∼100 kb) and largely revealed segments of local contact enrichment referred to as TADs (Dixon et al. 2012; Nora et al. 2012). Neighboring TADs show approximately two- to threefold enrichment of contacts within a domain as compared with interactions between neighboring domains. In contrast to compartments, they do not “checkerboard” with each other, and do not show specific enrichment of interactions when far away on the same or on different chromosomes (Mirny et al. 2019).
As the resolution and the depth of Hi-C data increased (Rao et al. 2014), new patterns such as focal enrichments of interactions between CTCF sites at TAD borders started to emerge. Commonly referred to as “loops,” such enrichments may be better called “dots” as they represent transient (Cattoni et al. 2017; Finn et al. 2019; Luppino et al. 2020; Su et al. 2020), rather than stable loops as was originally believed. Another frequent, near-diagonal pattern is a “stripe” or a “line” that emanates from a CTCF site (Fudenberg et al. 2016, 2017; Vian et al. 2018; Barrington et al. 2019), and reflects an enrichment of contacts between the CTCF and its neighborhood, extending sometimes for up to ∼1–3 Mb. Most recent and highest resolution Micro-C and Hi-C data revealed the abundance of such “dot” and “stripe” patterns, and that many stripes appear to include a series of dots (Hsieh et al. 2020; Krietenstein et al. 2020; Oksuz et al. 2020).
High-resolution microscopy extended our understanding of these patterns. First, microscopy has shown that maps of the median distance between loci closely resemble Hi-C maps (Bintu et al. 2018), with intra-TAD distances being smaller than inter-TAD ones (Finn et al. 2019). Second, and consistent with single-cell Hi-C (Nagano et al. 2013; Flyamer et al. 2017; Stevens et al. 2017), microscopy has observed a great deal of cell-to-cell variation in distances for pairs of loci within the same or in different TADs (Bintu et al. 2018; Finn et al. 2019; Su et al. 2020), further supporting the notion that TADs reflect enrichments of contacts rather than solid or even spatially discernable structures. Third, microscopy has clearly shown that focal enrichments of contacts between CTCF sites (“dots” or “loops”) do not represent stable loops, with their ends being in detectable proximity (<200–300 nm) in merely 10% of cells in the population at any given moment in time (Cattoni et al. 2017; Finn et al. 2019).
A surprising result of the last few years is that all these intricate patterns of contact enrichments are produced by a single process: an active (ATP-dependent) process of loop extrusion by cohesins that is occluded by extrusion barriers that are created largely by CTCF proteins (for review, see Fudenberg et al. 2017), and possibly modulated by interplay with other processes such as transcription.
THE MECHANICS OF LOOP EXTRUSION
Loop extrusion is a process whereby a molecular motor binds the chromatin fiber or DNA (e.g., in bacteria) and reels it in from one or both sides forming a progressively larger loop (Alipour and Marko 2012). Hypotheses about such a mechanism and its role in somatic recombination (Wood and Tonegawa 1983), enhancer–promoter interactions and chromosome organization during interphase (Riggs 1990), and DNA compaction and segregation during mitosis (Nasmyth 2001) were appearing in early literature, about once per decade, but remained largely unexplored until recently. The first computational models of loop extrusion suggested that it can generate arrays of nested or consecutive loops (Alipour and Marko 2012). Polymer models further showed that such arrays of loops can reproduce Hi-C data for mitotic chromosomes (Naumova et al. 2013). Polymer models further showed that extrusion can compact chromosomes and segregate sister chromatids (Goloborodko et al. 2016a,b), and help them disentangle topologically (Brahmachari and Marko 2019; Orlandini et al. 2019), thus suggesting a critical role of the loop extrusion process in mitosis.
Critically, simulations have shown that when extrusion is combined with barriers that can stop or pause it at specific genomic positions (Sanborn et al. 2015; Fudenberg et al. 2016), extrusion generates a broad range of patterns of local contact enrichment, such as TADs, dots, and stripes. Computational studies (Fudenberg et al. 2016) further hypothesized that SMC complexes (cohesins, condensins, etc.), believed at the time to be passive rings (Nasmyth and Haering 2009), are actually loop-extruding motors, and DNA-bound CTCF proteins are extrusion barriers. A broad range of experimental evidence from in vivo depletion of cohesin (Gassler et al. 2017; Haarhuis et al. 2017; Rao et al. 2017; Schwarzer et al. 2017; Wutz et al. 2017) and CTCF (Nora et al. 2017) to direct single-molecule visualizations (Lazar-Stefanita et al. 2017; Ganji et al. 2018; Davidson et al. 2019; Kim et al. 2019; Golfier et al. 2020; Kong et al. 2020; Cockram et al. 2021) support the loop extrusion mechanisms in eukaryotes.
Studies in bacteria, in turn, provided extensive evidence of loop extrusion by bacterial SMC complexes that juxtapose chromosomal arms (Wang et al. 2017; Gruber 2018; Böhm et al. 2020) and facilitate lengthwise compaction of chromosomes (Mäkelä and Sherratt 2020a,b). The presence of a specific loading (ParS) site for bacterial condensins (bSMC) in several bacteria lead to emergence of a distinct pattern on a Hi-C map, a secondary diagonal orthogonal to the main one (Gruber 2014; Wang et al. 2017; Böhm et al. 2020). Studies in bacteria further allowed (1) direction of extrusion in vivo and direct measurement of the extrusion speed (∼25 kb/min in Caulobacter [Tran et al. 2017] and ∼50 kb/min in Bacillus [Wang et al. 2017]); (2) characterizing specific loading and randomly loading of different extruding SMCs (Lioy et al. 2020; for review, see Gruber 2018); (3) measuring slow-down caused by interactions with transcriptional machinery (Brandão et al. 2019); and (4) genomic engineering experiments to create a “collision track” for examining interactions between SMCs (Brandão et al. 2021), suggesting that SMCs can bypass each other. Bypassing condensins has been observed in single-molecule experiments with yeast condensin on naked DNA (Kim et al. 2020).
Together Hi-C, microscopy, and single-molecule experiments draw the following picture of chromosome organization by loop extrusion in many eukaryotes including mammals:
By forming ∼100–200 kb loops, extrusion reduces contour length between every pair of loci, and hence increases the contact frequency between all regions on the same chromosome. This increase in contact frequency is most prominent for loci separated by <3 Mb as seen in Hi-C, and accompanied by reduced spatial distance as seen in superresolution microscopy (Bintu et al. 2018). Because loops are unlikely to get formed across TAD borders (depending on the permeability of CTCF barriers to cohesins), the contact frequency within TADs is increased more than between TADs.
Individual extruded loops have not been directly observed in vivo neither in microscopy nor in Hi-C because they are transient and dynamic (cohesin exchanges every 5–20 min [Gerlich et al. 2006a; Hansen et al. 2017; Wutz et al. 2017]); interactions they create can be practically indiscernible from myriads of other random interactions. Extruded loops, however, modify Hi-C maps in a predictable manner, so loops sizes (∼100–200 kb) can be inferred from the scaling P(s) curves of Hi-C data (Gassler et al. 2017).
Elevated contact frequency within a TAD and its insulation from neighboring TADs is caused by different positions of individual extruded loops in different cells and at different times, rather than caused by a fixed loop between two CTCF borders. Simulations show that a fixed loop does not increase contact frequency within a loop interior, nor insulates its interior from the flanks or neighboring loops (Fudenberg et al. 2016).
Similar to TADs, other local patterns such as dots and stripes do not reflect stable loops and constitute enrichments of contact frequency. They emerge as a result of pausing of extruding cohesin at specific genomic locations.
Functional Roles of Loop Extrusion
What possible functions can this versatile mechanism and intricate pattern of contact enrichments that it generates have? It is possible that during interphase the dynamic process of extrusion, with 5–20 min turnover time (Gerlich et al. 2006a; Hansen et al. 2017; Wutz et al. 2017), can be more important than the loops it produces (Liu et al. 2021). Extrusion can bring together distant genomic elements, such as enhancers and promoters, and do so more reliably than 3D search alone because it acts strictly in cis. Importantly, such extrusion-mediated contacts can be controlled by placing CTCF barriers between elements that should not interact. CTCF barriers can also facilitate contacts: When cohesin stalls on CTCF, it can continue reeling DNA in on the other side, thus scanning long genomic regions (Fudenberg et al. 2017). Such scanning is implicated in stochastic promoter choice in the protocadherin gene cluster (Guo et al. 2012; Canzio et al. 2019), and stochastic choice of partner sites for somatic recombination in the V(D)J locus (Ba et al. 2020). One possibility is that cohesin/CTCF-mediated scanning (Kraft et al. 2019) can also allow a locus (e.g., a promoter) to scan a long genomic region in search for its enhancer and/or integrating information from multiple scanned enhancers. Because CTCF bound sites constitute directional barriers (Rao et al. 2014; Vietri Rudan et al. 2015) (i.e., halt extrusion only if cohesin approaches CTCF from one side, but not the other) CTCF can direct scanning in a particular direction. Other mechanisms such as 3D spatial contacts and phase-separation/affinity-mediated contacts can be hardly controlled by insulators or established in a specific direction along the genome. Broadly, such a system of extruders and barriers opens many possibilities for controlled and targeted long-range communication along the genome.
Open questions:
Molecular mechanism of loop extrusion by SMCs remains mysterious. Recent cryo-electron microscopy (EM) structures (Hassler et al. 2019; Higashi et al. 2020; Lee et al. 2020; Li et al. 2020b; Muir et al. 2020; Shi et al. 2020) and single-molecule measurements (Ryu et al. 2020) inspired different molecular models (Marko et al. 2019; Higashi et al. 2021), but more detailed physical and structural characterization of SMC complexes at different steps of the extrusion cycle is essential for understanding this vital process.
Loop extrusion dynamics in live cells remains elusive. Despite the wealth of evidence from static perturbation experiments, direct visualization of loop extrusion in single-molecule experiments in vitro, and observation of extrusion in bacteria, dynamics of loop extrusion in live eukaryotic cells have not been captured. Although challenging because of fluctuations of live chromosomes, such experiments can provide direct evidence of extrusion in vivo. Moreover, in vivo characterization of extrusion dynamics (Brandão et al. 2021) could allow measuring the speed and the processivity of different SMC complexes, their interactions with CTCF and other factors, their activity through the cell cycle, and interactions with each other and with other processes on crowded chromatin templates and DNA.
Functional roles of the loop extension and patterns of contacts that it generates during interphase remain a subject of debate. On the one hand, loop extrusion is an attractive mechanism for mediating enhancer–promoter interactions because of its ability to facilitate contacts, and owing to control over direction and extent of such contacts exerted by CTCF and other extrusion barriers. On the other hand, depletion of cohesin and CTCF appear to be essential for transcriptional response to stimuli (Cuartero et al. 2018; Stik et al. 2020; Weiss et al. 2021), but dispensable for the maintenance of transcription of the majority of constitutive genes (Rao et al. 2017; Schwarzer et al. 2017).
What are the rules and regulation of SMC traffic along the genome? Understanding these rules is essential for understanding their possibly broad-range functional roles. These include understanding interactions between extruders when they meet each other, effects of positioned and random barriers (Dequeker et al. 2020) on extruders and vice versa. Moreover, it is crucial to understand how extrusion operates on crowded chromatin and interferes with and/or is modulated by other processes such as transcription of genic and nongenic regions, replication, condensate formation, and tethering of loci.
Are there other roles for loop extrusion? Other possible function of extrusion may be roles in facilitation of double-strand break (DSB) repair (Piazza et al. 2020; Arnould et al. 2021; Mirny 2021) and homology search on DNA damage and in meiosis (Patel et al. 2019; Schalbetter et al. 2019; Grey and de Massy 2021; Jin et al. 2021), its potential role in spreading of histone marks (Collins et al. 2020; Li et al. 2020a), etc.
TETHERING TO SUBNUCLEAR STRUCTURES
Besides loop extrusion and compartmentalization, a third mechanism of tethering can act on chromosomes to determine where loci are positioned with respect to other nuclear structures. Most prominently, loci can become tethered or anchored near the nuclear periphery. Early microscopic observations already established that densely staining heterochromatin is located at the nuclear periphery, and also around nucleoli. Genomic assays have been instrumental in identifying and characterizing chromosomal regions that are positioned at the nuclear periphery. The DamID method (van Steensel et al. 2001) has been used to label and then identify genomic loci (lamina-associated domains [LADs]) that are spatially proximal to the nuclear lamina. These studies found that loci near the lamina are repressed or expressed at very low levels (Pickersgill et al. 2006; Guelen et al. 2008). Chromatin in these domains is enriched for histone modifications such as H3K9me2 and H3K9me3, is late replicating, and is enriched in LINE elements (van Steensel and Belmont 2017). These are all features that are typical for inactive heterochromatin.
Following single cells through mitosis showed that loci that are located at the periphery in one cell cycle, can become repositioned in daughter cells away from the periphery, suggesting that nuclear positioning for these loci is not strictly heritable but more stochastic (Kind et al. 2013). Some LADs can be also localized around the nucleolus. This is confirmed by other experiments in which loci at or around the nucleolus were mapped by isolating nucleoli and analyzing the associated DNA (van Koningsbruggen et al. 2010). These studies identified a set of loci, nucleolus-associated domains (NADs) that have chromatin features that are similar to LADs (van Koningsbruggen et al. 2010; Vertii et al. 2019). More recent studies in mouse embryonic stem cells identified two types of NADs: one that resembles LADs (type I) and a second type (type II) that is characterized by higher levels of gene expression, the facultative heterochromatin mark H3K27me3, and early replication (Vertii et al. 2019). Thus, a subset of heterochromatic domains can be either tethered to the lamina or is associated with the nucleolus (and probably alternates between these locations in different cells in the population), whereas a second set of facultative heterochromatin is specifically localized at the nucleolus.
The mechanisms by which loci are tethered to either the nuclear periphery of the nucleolus are not fully understood in detail. Little is known about the factors involved in localizing chromatin domains near the nucleolus. Some factors have been identified to regulate or directly mediate the association of LADs with the nuclear periphery. Although LADs have been identified by their proximity to lamin B, lamins appear not directly required for association of LADs with the nuclear periphery at least in mouse embryonic stem cells (Amendola and van Steensel 2015). There are likely other factors that may contribute to the tethering of these domains. One candidate is the lamin B receptor (LBR). Studies in mouse cells showed that LBR acts sequentially with lamin A/C to tether heterochromatin (Solovei et al. 2013). When both factors are absent, heterochromatin localizes at the center of the nucleus with euchromatin at the periphery (see above). This unusual organization has been referred to as an inverted nucleus given that the positions of heterochromatin and euchromatin are reversed compared with their canonical organization in most cells. In natural settings, such inverted organization is observed in rod cells of nocturnal animals (Solovei et al. 2009). Other potential factors contributing to peripheral localization of heterochromatin include histone methyltransferases (e.g., MET-2 and SET-25 in Caenorhabditis elegans) (Towbin et al. 2012). Similarly, in mammalian cells, the histone methyltransferase G9a has been shown to regulate association of loci to the nuclear lamina (Kind et al. 2013).
Although tethering of heterochromatin is well-characterized, tethering of euchromatin is less understood. For instance, highly active loci can be localized at nuclear speckles (Zhang et al. 2021). Similarly, genes with many exons are enriched in contacts with other similar genes in cis and in trans, suggesting a role of splicing machinery in mediating such interactions. Whether this localization is caused by tethering of loci to these splicing factor-enriched nuclear bodies or whether speckles form around clusters of active loci is currently not known.
Open questions:
Why is heterochromatin tethered to the periphery? What are functional advantages of the conventional nuclear architecture with heterochromatin tethered to the lamina at the nuclear periphery? The inverted organization appears to be the default and to be compatible with gene expression.
Does tethering loci to the nuclear periphery help maintain transcriptional repression and repressive histone marks of heterochromatic regions? Does tethering of active regions to nuclear speckles play similar roles in maintaining transcription, its histone marks, and cotranscriptional splicing?
To what extent is chromatin tethered to intranuclear lamins? Lamins are found in the nucleoplasm but their roles are not well understood.
Does tethering of heterochromatin change mechanical properties to the nucleus? Mechanical properties of chromosomes and nuclei are increasingly understood to be important for cells within tissues and when cells are mobile. Does tethering reduce general mobility of chromatin and is this important for control of genomic processes such as transcription, replication, and repair?
TOPOLOGICAL EFFECTS
Topological effects do not constitute a mechanism of folding by themselves but they constrain and modulate what other mechanisms can achieve. Topological constraints prevent chromatin fibers from passing each other, unless topoisomerase II facilitates such passing. Activity of topoisomerase II during interphase is believed to be modest (Canela et al. 2019), arguing that topological constraints can play an important role in the shaping of interphase chromosomes. As such, topological effects can constrain and modulate chromosome folding driven by the three major mechanisms described above.
Topological constraints are known to have two major effects on polymer systems: (1) dramatic slowdown of equilibration, thus producing long-lived nonequilibrium states, and (2) generating of a different equilibrium state in a topologically constrained system (e.g., an unknotted ring that remains unknotted can behave differently than a ring in which strands can pass by each other) (Halverson et al. 2014).
Topological effects have been implicated in a range of chromosome phenomena. Formation of chromosomal territories have been attributed to formation of a nonequilibrium state after exit from mitosis (Abramo et al. 2019) when chromosomes do not have time to mix with each other (Rosa and Everaers 2008; Rosa et al. 2010). Formation of the largely unknotted fractal globule, as evident from Hi-C data (Lieberman-Aiden et al. 2009), is also attributed to a nonequilibrium state in which a polymer chain is crumpled attributed to topological interactions (Grosberg et al. 1988). Theoretically, such nonequilibrium states in which chains form territories and crumpled globules may nevertheless gradually, yet very slowly, equilibrate. Alternatively, nonconcatenated ring polymers form an equilibrium state with highly territorial rings, each forming a crumpled (fractal) globule (Halverson et al. 2014). Whether topological interactions lead to formation of long-lived nonequilibrium states or to equilibrium states remains to be understood. Recent studies that used modeling of Hi-C (Goundaroulis et al. 2020) and a complementary approach of multicontact 3C came to the conclusion that interphase chromatin is indeed largely unknotted (Tavares-Cadete et al. 2020). The interplay between topological constraints and major mechanisms discussed above are yet to be understood.
THE INTERPLAY OF MAJOR FOLDING MECHANISMS
Several aspects of interphase organization result from complex interplay of the three major mechanisms described above.
Extrusion versus Compartmentalization
One of the first successful cohesin and Wapl depletion studies (Haarhuis et al. 2017; Schwarzer et al. 2017) observed that, surprisingly, on depletion or enrichment of chromatin associated cohesin, not only cohesin-dependent features, but also compartmentalization was affected. Specifically, cohesin depletion resulted not only in the disappearance of extrusion-mediated features (TADs, dots, and stripes), but also in the strengthening of compartments (Schwarzer et al. 2017). An increase in cohesin residence time upon Wapl depletion, in turn, results in the weakening of compartmentalization and less-defined LADs (Haarhuis et al. 2017). These and the following studies suggested extensive interplay between loop extrusion and compartmentalization (Nuebler et al. 2018).
Cohesin-depleted chromosomes showed stronger and finer patterns of compartmentalization: Longer compartment regions got split into shorter regions. Interestingly, such finer compartments better reflect patterns of histone modification with H3K9me3- and H3K27me3-containing regions becoming shorter B compartment regions. Similarly, shorter H3K27ac islands become A compartmental regions. In wild-type, B and A regions are more stretched and less correlated with histone marks of repression and activity. These results suggested that cohesin depletion revealed “innate” compartmentalization preferences that were partially washed off by loop extrusion in the wild-type (Schwarzer et al. 2017). Simulations further showed that such masking of fine compartmentalization results from loop extrusion interfering with compartmental phase separation (Nuebler et al. 2018). Furthermore, simulations showed that increased cohesin residence time, as observed after Wapl depletion, leads to further weakening of compartmentalization.
The underlying physics of this interference is being actively explored. Polymer simulations in which chromosomes experience both compartmentalizing interactions (attraction between B regions) and active loop extrusion show that extrusion indeed weakens compartmentalization. Simulations further show that short compartmental regions are more sensitive to extrusion than long ones. Consistently, cohesin depletion experiments observed a similar phenomenon: small compartments are washed off in the wild-type while visible after cohesin depletion (Nuebler et al. 2018). Simulations of an increased extrusion activity reproduce phenomena observed in Wapl-depleted cells (Tedeschi et al. 2013; Haarhuis et al. 2017) (i.e., formation of overcompacted interphase [“vermicelli”] chromosomes accompanied by weakening of compartmentalization). Importantly, simulations show that reduction of compartmentalization by extrusion is not owing to loops per se, but rather owing to the active process of extrusion that “stirs” and mixes chromosomal regions, making it difficult for them to phase-separate (Nuebler et al. 2018).
Together, these results indicate that patterns of compartmentalization observed in wild-type interphase cells result from the interplay of phase separation of compartments and active loop extrusion.
Compartmentalization versus Tethering
Loss of tethering results in “nuclear inversion” in both natural systems such as the rods of nocturnal animals and in mutants that lack both LBR and lamin A/C (Solovei et al. 2013). In the inverted nucleus, heterochromatic regions occupy the center of the nucleus, while the euchromatic compartment is located at the nuclear periphery, thus inverting the conventional organization (Solovei et al. 2016). Hi-C and microscopy showed that despite inversion, A and B compartmental regions remain spatially segregated (Falk et al. 2019). Thus, compartmentalization by itself is independent of tethering of loci to the periphery. Polymer models showed that compartmentalization, when disentangled from tethering in inverted nuclei, requires strong attractive interactions between heterochromatic regions and weak, if any, attractions between euchromatic regions. Polymer simulations that combine mechanisms for compartmentalization and tethering of heterochromatin to the nuclear periphery are sufficient to explain the observed nuclear organization in typical mammalian cells (Falk et al. 2019b). Furthermore, restoration of lamin A/C activity in rod cells leads to partial de-inversion. Together, these results indicate that conventional nuclear organization results from the joined activity of compartmentalization and tethering.
Interplay between Transcription, Compartmentalization, and Loop Extruders
Transcription can sometimes play a dominant role in chromosome folding. For instance, domain formation in bacteria is reduced or absent when transcription is blocked (Le et al. 2013). Transcription and RNA processing machinery can also influence chromosome folding in more subtle ways by interfering with folding mechanisms described above (Hnisz et al. 2017; Hilbert et al. 2018; Trojanowski et al. 2021). This is an area of active research, and insights into this interplay can further our still limited understanding of how transcription affects chromosome folding and vice versa. For example, in mammalian cells, transcription can have effects on nuclear positioning and compartment association of loci. As mentioned above, highly expressed genes can cluster together especially when extensively spliced. This may be related to the observed positioning of active genes around nuclear speckles (Zhang et al. 2021). Large and highly expressed genes can become covered with nascent RNA–protein complexes and this results in stiffening of the chromatin fiber and the relocalization of the gene away from its chromosomal territory and into the nuclear center (Leidescher et al. 2020). It is important to point out that the overall effect of transcription on genome-wide chromosome organization can be quite subtle: Blocking transcription, or entirely removing RNA polymerases using inducible degron approaches has only minor effects on compartmentalization and other features per se (Vian et al. 2018; Barutcu et al. 2019; Jiang et al. 2020; Olan et al. 2020).
The RNA transcription machinery and the loop extrusion machinery also directly interact along chromosomes. For instance, early studies in budding yeast have shown that RNA polymerase can push cohesin rings toward sites of convergent transcription (Lengronne et al. 2004). Given that in Saccharomyces cerevisiae cohesin is located on chromosomes predominantly in the S phase and G2/M, this could be cohesive cohesin. In mammalian cells, RNA polymerase can modulate the position of cohesin, either by acting as a passive extrusion barrier or by actively pushing cohesin complexes toward 3′ ends of the active gene (Busslinger et al. 2017; Olan et al. 2020). In bacteria, polymerase can act as a moving barrier to extruding SMC complexes resulting in the slower extrusion against the direction of transcription (Brandão et al. 2019).
Open questions:
What other mechanisms shape chromosome organization and how do they interfere with these mechanisms? For example, can random and transient interactions (e.g., mediated by HP1-family proteins) lead to weak gelation of chromatin?
What is the impact of heterochromatic interactions and tethering on loop extrusion by cohesin and binding/activity of CTCF? Conversely, can extrusion interfere with tethering or maintenance of heterochromatin and associated histone marks?
What kinds of interplay between the various folding mechanisms and transcription can enhance or weaken chromosome territoriality? Does territoriality benefit from extrusion, tethering or compartmentalization? Conversely, can territoriality interfere or modulate compartmentalization?
DIFFERENTIAL IMPLEMENTATION OF THE SAME FOLDING MECHANISMS DRIVES CELL-CYCLE CHANGES IN CHROMOSOME SHAPE
The three mechanisms of chromosome folding described above can explain folding properties observed for chromosomes during interphase. However, chromosomes change their shape dramatically through the cell cycle. In mitosis, chromosomes form compacted rods with sister chromatids largely separated but running side-by-side. The large morphological differences between interphase and mitotic chromosomes suggest that different mechanisms may be involved in folding chromosomes during these different cell-cycle stages. However, it now appears that in many eukaryotes the same mechanisms operate, but performed by different SMC complexes (Dekker and Mirny 2016; Goloborodko et al. 2016a; Fudenberg et al. 2017).
As cells enter prophase, condensin II complexes take over the role of cohesin to form mitotic chromatin loops. In contrast to cohesin, the residence time of condensin II on chromatin is much longer (Gerlich et al. 2006b; Hansen et al. 2017), leading to more stable extruded loops. Tightly packed arrays of stable loops lead to rod-shaped mitotic chromosomes (Goloborodko et al. 2016a,b). Although during prophase, chromatin remains tethered to the periphery, as the nuclear envelope breaks down chromosomes are mostly untethered during prometaphase.
As cells exit mitosis, condensins become inactivated after the metaphase–anaphase transition, and during cytokinesis, cohesin takes over again as the main loop extrusion factor (Abramo et al. 2019; Zhang et al. 2019; Kang et al. 2020). The more transient nature of cohesin-mediated loops, and their lower density along the chromosome, leads to a more decondensed chromosome that may be sufficient to allow long-range compartmental interactions to reform. Chromosomes also become tethered again at the nuclear envelope during the subsequent G1 (van Schaik et al. 2020; Wong et al. 2020).
In the above picture, it is the alternation between cohesin-driven loop extrusion and condensin-driven loop extrusion that leads to chromosome morphologies that appear very different. Interestingly, in mutants that stabilize cohesin-mediated loops (e.g., by removal of the cohesin-unloading factor WAPL), interphase chromosomes form so-called vermicelli chromosomes that resemble prophase threads (Tedeschi et al. 2013). This supports the proposal that similar mechanisms are at play in interphase and mitosis but that they are implemented in quantitatively different ways.
Compartmentalization of chromosomes rapidly disappears as cells enter prophase. Whether the biophysical process of compartmentalization is still active in mitosis, but simply overridden by other processes is not known at this moment. For instance, the formation of relatively stiff rods could be sufficient to prevent long-range compartmental interactions. However, the observation that meiotic prophase chromosomes are elongated rods but still show compartmentalization by Hi-C would argue rod formation itself is not sufficient to erase compartments. Similarly, in vermicelli interphase chromosomes (in cells depleted for WAPL), the chromosomes form long threads but compartmentalization is still detectable (although reduced). Alternatively, the process of compartmentalization may be actively turned off during mitosis, and then turned on again after cells exit mitosis. One way compartmentalization could be turned off is by removing or inactivating key factors that facilitate phase separation by acting as bridging factors linking active or inactive chromatin domains. Such factors can include the histones themselves, or nonhistone factors that bind either active or inactive chromatin domains. During mitosis, several histone residues become phosphorylated and perhaps this leads to loss of attractions between chromatin domains. Alternatively, or in addition, it has been shown that phosphorylation of histone tail residues including H310 and H3T3 prevents binding of other factors such as heterochromatin protein 1 (HP1) (Fischle et al. 2005; Hirota et al. 2005) or TFIID (Varier et al. 2010), respectively, which may in turn lead to loss of compartmentalization.
Open questions:
How is compartmentalization modulated during the cell cycle, differentiation, and aging? Compartmentalization appears highly dynamic during the cell cycle, cell-state transitions, and as cells age. Is this a regulated process? If so, how is it regulated?
What is the interplay between cohesin and condensin during prophase and prometaphase? During prophase, both cohesins and condensins are acting along chromosomes. Is there interference or collaboration between the complexes and how does that affect folding and segregating chromosomes during mitosis?
Is there an interplay between condensins and cohesins during mitotic exit? During mitotic exit, condensin inactivation and cohesin reloading appear to be temporally separated (Abramo et al. 2019). Is this a regulated process to avoid interference between these folding machines? If so, how are these processes coordinated?
CELL-TYPE-SPECIFIC CHROMOSOME FOLDING AND NUCLEAR ORGANIZATION
In different cell types, different parts of the genome are active or repressed. For instance, in each cell type specific genes, regulatory elements, and other functional elements such as CTCF sites and enhancers are active. The same folding mechanisms described above will act along chromosomes leading to cell-type-specific patterns of chromosome structures. For instance, in most cell types, compartmentalization of active and inactive chromatin domains is observed, but given that different loci are active and inactive dependent on the cell type, the composition of the A and B compartments is different as well. Similarly, cohesin-mediated loop extrusion will occur in most cell types, but the positions in which cohesin is loaded, and the genomic position of blocks to extrusion, most notably CTCF-bound sites, can differ between cell types. Finally, promoter–enhancer interactions that could be mediated by both loop extrusion or phase separation will differ between cell types given that different promoters and enhancers are active.
Interestingly, there are examples of cell types that display unique global nuclear organizations because specific mechanisms of chromosome folding themselves have been altered. One example is that of cells with inverted nuclei (see above). In rod photoreceptor cells of nocturnal mammals such as mice, the nucleus is inverted so that all heterochromatic loci are located in the center with active chromatin located peripherally (Solovei et al. 2009). Such inverted organization changes the optical properties of the nucleus and this is beneficial for photodetection. As mentioned above, this organization arises when the tethering of heterochromatin to the periphery is turned off.
Pro-B and pre-B cells represent another example of cells in which global chromosome organization is altered by modulating a specific mechanism of nuclear organization, in this case by changing cohesin-mediated loop extrusion. In these cells, Pax5 represses the expression of the cohesin-unloading factor WAPL. As a result, cohesin is more stably associated with chromatin and generates larger and more stable loops genome-wide (Hill et al. 2020). In pro-B and pre-B cells, this is thought to be important to facilitate the very long-range interactions required for contraction of the 2.8-Mb-long immunoglobulin heavy chain (Igh) locus during V(D)J recombination.
Open questions:
What other mechanisms drive chromosome folding and nuclear organization? Some cell types display unique chromosome conformations that could be formed by additional mechanisms. For instance, in olfactory neurons especially dense heterochromatic clusters are formed in the central part of the nucleus.
How malleable and/or reversible is genome and nuclear organization (e.g., during reprogramming, senescence, aging, and disease)?
ACKNOWLEDGMENTS
We are grateful to members of Mirny and Dekker laboratories for many discussions of these mechanisms and chromosome phenomena, to Irina Solovei for deep knowledge of the field she shared with us and for microscopy images of conventional and inverted nuclei, and to George Spracklin for producing DamID/Protect-seq/H3K9me3 panel of Figure 1. L.M. and J.D. are supported by a grant from the National Institutes of Health Common Fund 4D Nucleome Program (U54-DK107980, UM1-HG011536) and a grant from the National Human Genome Research Institute (NHGRI) to J.D. (HG003143). J.D. is an investigator of the Howard Hughes Medical Institute. L.M. is Blaize Pascal Chair of Ile-de-France, visiting Institut Curie.
Footnotes
Editors: Ana Pombo, Martin W. Hetzer, and Tom Misteli
Additional Perspectives on The Nucleus available at www.cshperspectives.org
REFERENCES
- Abramo K, Valton AL, Venev SV, Ozadam H, Fox AN, Dekker J. 2019. A chromosome folding intermediate at the condensin-to-cohesin transition during telophase. Nat Cell Biol 21: 1393–1402. 10.1038/s41556-019-0406-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alipour E, Marko JF. 2012. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res 40: 11202–11212. 10.1093/nar/gks925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amendola M, van Steensel B. 2015. Nuclear lamins are not required for lamina-associated domain organization in mouse embryonic stem cells. EMBO Rep 16: 610–617. 10.15252/embr.201439789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnould C, Rocher V, Finoux AL, Clouaire T, Li K, Zhou F, Caron P, Mangeot PE, Ricci EP, Mourad R, et al. 2021. Loop extrusion as a mechanism for formation of DNA damage repair foci. Nature 590: 660–665. 10.1038/s41586-021-03193-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ba Z, Lou J, Ye AY, Dai HQ, Dring EW, Lin SG, Jain S, Kyritsis N, Kieffer-Kwon KR, Casellas R, et al. 2020. CTCF orchestrates long-range cohesin-driven V(D)J recombinational scanning. Nature 586: 305–310. 10.1038/s41586-020-2578-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri M, Chotalia M, Fraser J, Lavitas LM, Dostie J, Pombo A, Nicodemi M. 2012. Complexity of chromatin folding is captured by the strings and binders switch model. Proc Natl Acad Sci 109: 16173–16178. 10.1073/pnas.1204799109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrington C, Georgopoulou D, Pezic D, Varsally W, Herrero J, Hadjur S. 2019. Enhancer accessibility and CTCF occupancy underlie asymmetric TAD architecture and cell type specific genome topology. Nat Commun 10: 2908. 10.1038/s41467-019-10725-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barutcu AR, Blencowe BJ, Rinn JL. 2019. Differential contribution of steady-state RNA and active transcription in chromatin organization. EMBO Rep 20: e48068. 10.15252/embr.201948068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu B, Mateo LJ, Su JH, Sinnott-Armstrong NA, Parker M, Kinrot S, Yamaya K, Boettiger AN, Zhuang X. 2018. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 362: eaau1783. 10.1126/science.aau1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm K, Giacomelli G, Schmidt A, Imhof A, Koszul R, Marbouty M, Bramkamp M. 2020. Chromosome organization by a conserved condensin-ParB system in the actinobacterium Corynebacterium glutamicum. Nat Commun 11: 1485. 10.1038/s41467-020-15238-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Mendelson Cohen N, Szabo Q, Fritsch L, Papadopoulos GL, Lubling Y, Xu X, Lv X, Hugnot JP, Tanay A, et al. 2017. Multiscale 3D genome rewiring during mouse neural development. Cell 171: 557–572.e24. 10.1016/j.cell.2017.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle S, Flyamer IM, Williamson I, Sengupta D, Bickmore WA, Illingworth RS. 2020. A central role for canonical PRC1 in shaping the 3D nuclear landscape. Genes Dev 34: 931–949. 10.1101/gad.336487.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmachari S, Marko JF. 2019. Chromosome disentanglement driven via optimal compaction of loop-extruded brush structures. Proc Natl Acad Sci 116: 24956–24965. 10.1073/pnas.1906355116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandão HB, Paul P, van den Berg AA, Rudner DZ, Wang X, Mirny LA. 2019. RNA polymerases as moving barriers to condensin loop extrusion. Proc Natl Acad Sci 116: 20489–20499. 10.1073/pnas.1907009116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandão HB, Ren Z, Karaboja X, Mirny LA, Wang X. 2021. DNA-loop-extruding SMC complexes can traverse one another in vivo. Nat Struct Mol Biol 10.1038/s41594-021-00626-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandão HB, Gabriele M, Hansen AS. 2021. Tracking and interpreting long-range chromatin interactions with super-resolution live-cell imaging. Curr Opin Cell Biol 70: 18–26. 10.1016/j.ceb.2020.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busslinger GA, Stocsits RR, van der Lelij P, Axelsson E, Tedeschi A, Galjart N, Peters JM. 2017. Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544: 503–507. 10.1038/nature22063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A, Maman Y, Huang SYN, Wutz G, Tang W, Zagnoli-Vieira G, Callen E, Wong N, Day A, Peters JM, et al. 2019. Topoisomerase II-induced chromosome breakage and translocation is determined by chromosome architecture and transcriptional activity. Mol Cell 75: 252–266.e8. 10.1016/j.molcel.2019.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canzio D, Nwakeze CL, Horta A, Rajkumar SM, Coffey EL, Duffy EE, Duffié R, Monahan K, O'Keeffe S, Simon MD, et al. 2019. Antisense lncRNA transcription mediates DNA demethylation to drive stochastic protocadherin α promoter choice. Cell 177: 639–653.e15. 10.1016/j.cell.2019.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoni DI, Cardozo Gizzi AM, Georgieva M, Di Stefano M, Valeri A, Chamousset D, Houbron C, Déjardin S, Fiche JB, González I, et al. 2017. Single-cell absolute contact probability detection reveals chromosomes are organized by multiple low-frequency yet specific interactions. Nat Commun 8: 1753. 10.1038/s41467-017-01962-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockram C, Thierry A, Gorlas A, Lestini R, Koszul R. 2021. Euryarchaeal genomes are folded into SMC-dependent loops and domains, but lack transcription-mediated compartmentalization. Mol Cell 81: 459–472.e10. 10.1016/j.molcel.2020.12.013 [DOI] [PubMed] [Google Scholar]
- Collins PL, Purman C, Porter SI, Nganga V, Saini A, Hayer KE, Gurewitz GL, Sleckman BP, Bednarski JJ, Bassing CH, et al. 2020. DNA double-strand breaks induce H2Ax phosphorylation domains in a contact-dependent manner. Nat Commun 11: 3158. 10.1038/s41467-020-16926-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuartero S, Weiss FD, Dharmalingam G, Guo Y, Ing-Simmons E, Masella S, Robles-Rebollo I, Xiao X, Wang YF, Barozzi I, et al. 2018. Control of inducible gene expression links cohesin to hematopoietic progenitor self-renewal and differentiation. Nat Immunol 19: 932–941. 10.1038/s41590-018-0184-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson IF, Bauer B, Goetz D, Tang W, Wutz G, Peters JM. 2019. DNA loop extrusion by human cohesin. Science 366: 1338–1345. 10.1126/science.aaz3418 [DOI] [PubMed] [Google Scholar]
- Dekker J, Mirny L. 2016. The 3D genome as moderator of chromosomal communication. Cell 164: 1110–1121. 10.1016/j.cell.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dequeker BJH, Brandão HB, Scherr MJ, Gassler J, Powell S, Gaspar I, Flyamer IM, Tang W, Stocsits R, Davidson IF, et al. 2020. MCM complexes are barriers that restrict cohesin-mediated loop extrusion. bioRxiv; 10.1101/2020.10.15.340356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dileep V, Ay F, Sima J, Vera DL, Noble WS, Gilbert DM. 2015. Topologically associating domains and their long-range contacts are established during early G1 coincident with the establishment of the replication-timing program. Genome Res 25: 1104–1113. 10.1101/gr.183699.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pierro M, Zhang B, Aiden EL, Wolynes PG, Onuchic JN. 2016. Transferable model for chromosome architecture. Proc Natl Acad Sci 113: 12168–12173. 10.1073/pnas.1613607113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zheng H, Huang B, Ma R, Wu J, Zhang X, He J, Xiang Y, Wang Q, Li Y, et al. 2017. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 547: 232–235. 10.1038/nature23263 [DOI] [PubMed] [Google Scholar]
- Falk M, Feodorova Y, Naumova N, Imakaev M, Lajoie BR, Leonhardt H, Joffe B, Dekker J, Fudenberg G, Solovei I, et al. 2019. Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature 570: 395–399. 10.1038/s41586-019-1275-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn EH, Pegoraro G, Brandão HB, Valton AL, Oomen ME, Dekker J, Mirny L, Misteli T. 2019. Extensive heterogeneity and intrinsic variation in spatial genome organization. Cell 176: 1502–1515.e10. 10.1016/j.cell.2019.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. 2005. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438: 1116–1122. 10.1038/nature04219 [DOI] [PubMed] [Google Scholar]
- Flyamer IM, Gassler J, Imakaev M, Brandão HB, Ulianov SV, Abdennur N, Razin SV, Mirny LA, Tachibana-Konwalski K. 2017. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544: 110–114. 10.1038/nature21711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. 2016. Formation of chromosomal domains by loop extrusion. Cell Rep 15: 2038–2049. 10.1016/j.celrep.2016.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G, Abdennur N, Imakaev M, Goloborodko A, Mirny LA. 2017. Emerging evidence of chromosome folding by loop extrusion. Cold Spring Harb Symp Quant Biol 82: 45–55. 10.1101/sqb.2017.82.034710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganji M, Shaltiel IA, Bisht S, Kim E, Kalichava A, Haering CH, Dekker C. 2018. Real-time imaging of DNA loop extrusion by condensin. Science 360: 102–105. 10.1126/science.aar7831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassler J, Brandão HB, Imakaev M, Flyamer IM, Ladstätter S, Bickmore WA, Peters JM, Mirny LA, Tachibana K. 2017. A mechanism of cohesin-dependent loop extrusion organizes zygotic genome architecture. EMBO J 36: 3600–3618. 10.15252/embj.201798083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J. 2006a. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr Biol 16: 1571–1578. 10.1016/j.cub.2006.06.068 [DOI] [PubMed] [Google Scholar]
- Gerlich D, Hirota T, Koch B, Peters JM, Ellenberg J. 2006b. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr Biol 16: 333–344. 10.1016/j.cub.2005.12.040 [DOI] [PubMed] [Google Scholar]
- Gibcus JH, Samejima K, Goloborodko A, Samejima I, Naumova N, Nuebler J, Kanemaki MT, Xie L, Paulson JR, Earnshaw WC, et al. 2018. A pathway for mitotic chromosome formation. Science 359: eaao6135. 10.1126/science.aao6135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girelli G, Custodio J, Kallas T, Agostini F, Wernersson E, Spanjaard B, Mota A, Kolbeinsdottir S, Gelali E, Crosetto N, et al. 2020. GPSeq reveals the radial organization of chromatin in the cell nucleus. Nat Biotechnol 38: 1184–1193. 10.1038/s41587-020-0519-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golfier S, Quail T, Kimura H, Brugués J. 2020. Cohesin and condensin extrude DNA loops in a cell cycle-dependent manner. eLife 9: e53885. 10.7554/eLife.53885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloborodko A, Imakaev MV, Marko JF, Mirny L. 2016a. Compaction and segregation of sister chromatids via active loop extrusion. eLife 5: e14864. 10.7554/eLife.14864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloborodko A, Marko JF, Mirny LA. 2016b. Chromosome compaction by active loop extrusion. Biophys J 110: 2162–2168. 10.1016/j.bpj.2016.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goundaroulis D, Lieberman Aiden E, Stasiak A. 2020. Chromatin is frequently unknotted at the megabase scale. Biophys J 118: 2268–2279. 10.1016/j.bpj.2019.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey C, de Massy B. 2021. Chromosome organization in early meiotic prophase. Front Cell Dev Biol 9: 688878. 10.3389/fcell.2021.688878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosberg AY, Nechaev SK, Shakhnovich EI. 1988. The role of topological constraints in the kinetics of collapse of macromolecules. J Phys (France) 49: 2095–2100. 10.1051/jphys:0198800490120209500 [DOI] [Google Scholar]
- Gruber S. 2014. Multilayer chromosome organization through DNA bending, bridging and extrusion. Curr Opin Microbiol 22: 102–110. 10.1016/j.mib.2014.09.018 [DOI] [PubMed] [Google Scholar]
- Gruber S. 2018. SMC complexes sweeping through the chromosome: going with the flow and against the tide. Curr Opin Microbiol 42: 96–103. 10.1016/j.mib.2017.10.004 [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951. 10.1038/nature06947 [DOI] [PubMed] [Google Scholar]
- Guo Y, Monahan K, Wu H, Gertz J, Varley KE, Li W, Myers RM, Maniatis T, Wu Q. 2012. CTCF/cohesin-mediated DNA looping is required for protocadherin α promoter choice. Proc Natl Acad Sci 109: 21081–21086. 10.1073/pnas.1219280110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarhuis JHI, van der Weide RH, Blomen VA, Yáñez-Cuna JO, Amendola M, van Ruiten MS, Krijger PHL, Teunissen H, Medema RH, van Steensel B, et al. 2017. The cohesin release factor WAPL restricts chromatin loop extension. Cell 169: 693–707.e14. 10.1016/j.cell.2017.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halverson JD, Smrek J, Kremer K, Grosberg AY. 2014. From a melt of rings to chromosome territories: the role of topological constraints in genome folding. Rep Prog Phys 77: 022601. 10.1088/0034-4885/77/2/022601 [DOI] [PubMed] [Google Scholar]
- Hansen AS, Pustova I, Cattoglio C, Tjian R, Darzacq X. 2017. CTCF and cohesin regulate chromatin loop stability with distinct dynamics. eLife 6: e25776. 10.7554/eLife.25776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassler M, Shaltiel IA, Kschonsak M, Simon B, Merkel F, Thärichen L, Bailey HJ, Macošek J, Bravo S, Metz J, et al. 2019. Structural basis of an asymmetric condensin ATPase cycle. Mol Cell 74: 1175–1188.e9. 10.1016/j.molcel.2019.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi TL, Eickhoff P, Simoes JS, Locke J, Nans A, Flynn HR, Snijders AP, Papageorgiou G, O'Reilly N, Chen ZA, et al. 2020. A structure-based mechanism for DNA entry into the cohesin ring. Mol Cell 79: 917–933.e9. 10.1016/j.molcel.2020.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi TL, Pobegalov G, Tang M, Molodtsov MI, Uhlmann F. 2021. A Brownian ratchet model for DNA loop extrusion by the cohesin complex. eLife 10: e67530. 10.7554/eLife.67530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbert L, Sato Y, Kimura H, Jülicher F, Honigmann A. 2021. Transcription organizes euchromatin similar to an active microemulsion. Nat Commun 12: 1360. 10.1038/s41467-021-21589-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand EM, Dekker J. 2020. Mechanisms and functions of chromosome compartmentalization. Trends Biochem Sci 45: 385–396. 10.1016/j.tibs.2020.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill L, Ebert A, Jaritz M, Wutz G, Nagasaka K, Tagoh H, Kostanova-Poliakova D, Schindler K, Sun Q, Bönelt P, et al. 2020. Wapl repression by Pax5 promotes V gene recombination by Igh loop extrusion. Nature 584: 142–147. 10.1038/s41586-020-2454-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Lipp JJ, Toh BH, Peters JM. 2005. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438: 1176–1180. 10.1038/nature04254 [DOI] [PubMed] [Google Scholar]
- Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. 2017. A phase separation model for transcriptional control. Cell 169: 13–23. 10.1016/j.cell.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh THS, Cattoglio C, Slobodyanyuk E, Hansen AS, Rando OJ, Tjian R, Darzacq X. 2020. Resolving the 3D landscape of transcription-linked mammalian chromatin folding. Mol Cell 78: 539–553.e8. 10.1016/j.molcel.2020.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug CB, Grimaldi AG, Kruse K, Vaquerizas JM. 2017. Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell 169: 216–228.e19. 10.1016/j.cell.2017.03.024 [DOI] [PubMed] [Google Scholar]
- Imakaev M, Fudenberg G, McCord RP, Naumova N, Goloborodko A, Lajoie BR, Dekker J, Mirny LA. 2012. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods 9: 999–1003. 10.1038/nmeth.2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerabek H, Heermann DW. 2012. Expression-dependent folding of interphase chromatin. PLoS ONE 7: e37525. 10.1371/journal.pone.0037525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Huang J, Lun K, Li B, Zheng H, Li Y, Zhou R, Duan W, Wang C, Feng Y, et al. 2020. Genome-wide analyses of chromatin interactions after the loss of Pol I, Pol II, and Pol III. Genome Biol 21: 158. 10.1186/s13059-020-02067-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Fudenberg G, Pollard KS. 2021. Genome-wide variability in recombination activity is associated with meiotic chromatin organization. Genome Res 10.1101/gr.275358.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone SE, Reyes A, Qi Y, Adriaens C, Hegazi E, Pelka K, Chen JH, Zou LS, Drier Y, Hecht V, et al. 2020. Large-scale topological changes restrain malignant progression in colorectal cancer. Cell 182: 1474–1489.e23. 10.1016/j.cell.2020.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost D, Carrivain P, Cavalli G, Vaillant C. 2014. Modeling epigenome folding: formation and dynamics of topologically associated chromatin domains. Nucleic Acids Res 42: 9553–9561. 10.1093/nar/gku698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Shokhirev MN, Xu Z, Chandran S, Dixon JR, Hetzer MW. 2020. Dynamic regulation of histone modifications and long-range chromosomal interactions during postmitotic transcriptional reactivation. Genes Dev 34: 913–930. 10.1101/gad.335794.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y, Xu Y, Chen X, Feng S, Liu Z, Sun Y, Yao X, Li F, Zhu W, Gao L, et al. 2017. 3D chromatin structures of mature gametes and structural reprogramming during mammalian embryogenesis. Cell 170: 367–381.e20. 10.1016/j.cell.2017.06.029 [DOI] [PubMed] [Google Scholar]
- Kerpedjiev P, Abdennur N, Lekschas F, McCallum C, Dinkla K, Strobelt H, Luber JM, Ouellette SB, Azhir A, Kumar N, et al. 2018. HiGlass: web-based visual exploration and analysis of genome interaction maps. Genome Biol 19: 125. 10.1186/s13059-018-1486-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Shi Z, Zhang H, Finkelstein IJ, Yu H. 2019. Human cohesin compacts DNA by loop extrusion. Science 366: 1345–1349. 10.1126/science.aaz4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Kerssemakers J, Shaltiel IA, Haering CH, Dekker C. 2020. DNA-loop extruding condensin complexes can traverse one another. Nature 579: 438–442. 10.1038/s41586-020-2067-5 [DOI] [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. 2013. Single-cell dynamics of genome-nuclear lamina interactions. Cell 153: 178–192. 10.1016/j.cell.2013.02.028 [DOI] [PubMed] [Google Scholar]
- Kong M, Cutts EE, Pan D, Beuron F, Kaliyappan T, Xue C, Morris EP, Musacchio A, Vannini A, Greene EC. 2020. Human condensin I and II drive extensive ATP-dependent compaction of nucleosome-bound DNA. Mol Cell 79: 99–111.e9. 10.1016/j.molcel.2020.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft K, Magg A, Heinrich V, Riemenschneider C, Schöpflin R, Markowski J, Ibrahim DM, Acuna-Hidalgo R, Despang A, Andrey G, et al. 2019. Serial genomic inversions induce tissue-specific architectural stripes, gene misexpression and congenital malformations. Nat Cell Biol 21: 305–310. 10.1038/s41556-019-0273-x [DOI] [PubMed] [Google Scholar]
- Krietenstein N, Abraham S, Venev SV, Abdennur N, Gibcus J, Hsieh THS, Parsi KM, Yang L, Maehr R, Mirny LA, et al. 2020. Ultrastructural details of mammalian chromosome architecture. Mol Cell 78: 554–565.e7. 10.1016/j.molcel.2020.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar-Stefanita L, Scolari VF, Mercy G, Muller H, Guérin TM, Thierry A, Mozziconacci J, Koszul R. 2017. Cohesins and condensins orchestrate the 4D dynamics of yeast chromosomes during the cell cycle. EMBO J 36: 2684–2697. 10.15252/embj.201797342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TBK, Imakaev MV, Mirny LA, Laub MT. 2013. High-resolution mapping of the spatial organization of a bacterial chromosome. Science 342: 731–734. 10.1126/science.1242059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BG, Merkel F, Allegretti M, Hassler M, Cawood C, Lecomte L, O'Reilly FJ, Sinn LR, Gutierrez-Escribano P, Kschonsak M, et al. 2020. Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism. Nat Struct Mol Biol 27: 743–751. 10.1038/s41594-020-0457-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidescher S, Ribisel J, Ullrich S, Feodorova Y, Hildebrand E, Bultmann S, Link S, Thanisch K, Mulholland C, Dekker J, et al. 2020. Spatial organization of transcribed eukaryotic genes. bioRxiv 10.1101/2020.05.20.106591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F. 2004. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430: 573–578. 10.1038/nature02742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Bronk G, Kondev J, Haber JE. 2020a. Yeast ATM and ATR kinases use different mechanisms to spread histone H2A phosphorylation around a DNA double-strand break. Proc Natl Acad Sci 117: 21354–21363. 10.1073/pnas.2002126117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Haarhuis JHI, Cacciatore ÁS, Oldenkamp R, van Ruiten MS, Willems L, Teunissen H, Muir KW, de Wit E, Rowland BD, et al. 2020b. The structural basis for cohesin-CTCF-anchored loops. Nature 578: 472–476. 10.1038/s41586-019-1910-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lioy VS, Junier I, Lagage V, Vallet I, Boccard F. 2020. Distinct activities of bacterial condensins for chromosome management in Pseudomonas aeruginosa. Cell Rep 33: 108344. 10.1016/j.celrep.2020.108344 [DOI] [PubMed] [Google Scholar]
- Liu NQ, Maresca M, van den Brand T, Braccioli L, Schijns MMGA, Teunissen H, Bruneau BG, Nora EP, de Wit E. 2021. WAPL maintains a cohesin loading cycle to preserve cell-type-specific distal gene regulation. Nat Genet 53: 100–109. 10.1038/s41588-020-00744-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luppino JM, Park DS, Nguyen SC, Lan Y, Xu Z, Yunker R, Joyce EF. 2020. Cohesin promotes stochastic domain intermingling to ensure proper regulation of boundary-proximal genes. Nat Genet 52: 840–848. 10.1038/s41588-020-0647-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson Q, Beltran B, Spakowitz AJ. 2018. Bottom-up modeling of chromatin segregation due to epigenetic modifications. Proc Natl Acad Sci 115: 12739–12744. 10.1073/pnas.1812268115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkelä J, Sherratt DJ. 2020a. Organization of the Escherichia coli chromosome by a MukBEF axial core. Mol Cell 78: 250–260.e5. 10.1016/j.molcel.2020.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkelä J, Sherratt D. 2020b. SMC complexes organize the bacterial chromosome by lengthwise compaction. Curr Genet 66: 895–899. 10.1007/s00294-020-01076-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko JF, De Los Rios P, Barducci A, Gruber S. 2019. DNA-segment-capture model for loop extrusion by structural maintenance of chromosome (SMC) protein complexes. Nucleic Acids Res 47: 6956–6972. 10.1093/nar/gkz497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirny LA. 2021. Cells use loop extrusion to weave and tie the genome. Nature 590: 554–555. 10.1038/d41586-021-00351-1 [DOI] [PubMed] [Google Scholar]
- Mirny LA, Imakaev M, Abdennur N. 2019. Two major mechanisms of chromosome organization. Curr Opin Cell Biol 58: 142–152. 10.1016/j.ceb.2019.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir KW, Li Y, Weis F, Panne D. 2020. The structure of the cohesin ATPase elucidates the mechanism of SMC-kleisin ring opening. Nat Struct Mol Biol 27: 233–239. 10.1038/s41594-020-0379-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. 2013. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502: 59–64. 10.1038/nature12593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. 2001. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet 35: 673–745. 10.1146/annurev.genet.35.102401.091334 [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. 2009. Cohesin: its roles and mechanisms. Annu Rev Genet 43: 525–558. 10.1146/annurev-genet-102108-134233 [DOI] [PubMed] [Google Scholar]
- Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J, 2013. Organization of the mitotic chromosome. Science 342: 948–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. 10.1038/nature11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG. 2017. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169: 930–944.e22. 10.1016/j.cell.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuebler J, Fudenberg G, Imakaev M, Abdennur N, Mirny LA. 2018. Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc Natl Acad Sci 115: E6697–E6706. 10.1073/pnas.1717730115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksuz BA, Yang L, Abraham S, Venev SV, Krietenstein N, Parsi KM, Ozadam H, Oomen ME, Nand A, Mao H, et al. 2020. Systematic evaluation of chromosome conformation capture assays. bioRxiv 10.1101/2020.12.26.424448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olan I, Parry AJ, Schoenfelder S, Narita M, Ito Y, Chan ASL, Slater GSC, Bihary D, Bando M, Shirahige K, et al. 2020. Transcription-dependent cohesin repositioning rewires chromatin loops in cellular senescence. Nat Commun 11: 6049. 10.1038/s41467-020-19878-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandini E, Marenduzzo D, Michieletto D. 2019. Synergy of topoisomerase and structural-maintenance-of-chromosomes proteins creates a universal pathway to simplify genome topology. Proc Natl Acad Sci 116: 8149–8154. 10.1073/pnas.1815394116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel L, Kang R, Rosenberg SC, Qiu Y, Raviram R, Chee S, Hu R, Ren B, Cole F, Corbett KD. 2019. Dynamic reorganization of the genome shapes the recombination landscape in meiotic prophase. Nat Struct Mol Biol 26: 164–174. 10.1038/s41594-019-0187-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. 2006. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet 38: 1005–1014. 10.1038/ng1852 [DOI] [PubMed] [Google Scholar]
- Ramani V, Deng X, Qiu R, Gunderson KL, Steemers FJ, Disteche CM, Noble WS, Duan Z, Shendure J. 2017. Massively multiplex single-cell Hi-C. Nat Methods 14: 263–266. 10.1038/nmeth.4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huang SC, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon KR, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, et al. 2017. Cohesin loss eliminates all loop domains. Cell 171: 305–320.e24. 10.1016/j.cell.2017.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs AD. 1990. DNA methylation and late replication probably aid cell memory, and type I DNA reeling could aid chromosome folding and enhancer function. Philos Trans R Soc Lond B Biol Sci 326: 285–297. 10.1098/rstb.1990.0012 [DOI] [PubMed] [Google Scholar]
- Rosa A, Everaers R. 2008. Structure and dynamics of interphase chromosomes. PLoS Comput Biol 4: e1000153. 10.1371/journal.pcbi.1000153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Becker NB, Everaers R. 2010. Looping probabilities in model interphase chromosomes. Biophys J 98: 2410–2419. 10.1016/j.bpj.2010.01.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein M, Colby R. 2003. Polymer physics. Oxford University Press, Oxford. [Google Scholar]
- Ryu JK, Rah SH, Janissen R, Kerssemakers JWJ, Dekker C. 2020. Resolving the step size in condensin-driven DNA loop extrusion identifies ATP binding as the step-generating process. bioRxiv 10.1101/2020.11.04.368506 [DOI] [Google Scholar]
- Sanborn AL, Rao SSP, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. 2015. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci 112: E6456–E6465. 10.1073/pnas.1518552112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalbetter SA, Fudenberg G, Baxter J, Pollard KS, Neale MJ. 2019. Principles of meiotic chromosome assembly revealed in S. cerevisiae. Nat Commun 10: 4795. 10.1038/s41467-019-12629-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, Fonseca NA, Huber W, Haering CH, Mirny L, et al. 2017. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551: 51–56. 10.1038/nature24281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Gao H, Bai XC, Yu H. 2020. Cryo-EM structure of the human cohesin-NIPBL-DNA complex. Science 368: 1454–1459. 10.1126/science.abb0981 [DOI] [PubMed] [Google Scholar]
- Shintomi K, Hirano T. 2011. The relative ratio of condensin I to II determines chromosome shapes. Genes Dev 25: 1464–1469. 10.1101/gad.2060311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. 2006. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet 38: 1348–1354. 10.1038/ng1896 [DOI] [PubMed] [Google Scholar]
- Solovei I, Kreysing M, Lanctôt C, Kösem S, Peichl L, Cremer T, Guck J, Joffe B. 2009. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137: 356–368. 10.1016/j.cell.2009.01.052 [DOI] [PubMed] [Google Scholar]
- Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, et al. 2013. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152: 584–598. 10.1016/j.cell.2013.01.009 [DOI] [PubMed] [Google Scholar]
- Solovei I, Thanisch K, Feodorova Y. 2016. How to rule the nucleus: divide et impera. Curr Opin Cell Biol 40: 47–59. 10.1016/j.ceb.2016.02.014 [DOI] [PubMed] [Google Scholar]
- Spracklin G, Pradhan S. 2020. Protect-seq: genome-wide profiling of nuclease inaccessible domains reveals physical properties of chromatin. Nucleic Acids Res 48: e16. 10.1093/nar/gkz1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens TJ, Lando D, Basu S, Atkinson LP, Cao Y, Lee SF, Leeb M, Wohlfahrt KJ, Boucher W, O'Shaughnessy-Kirwan A, et al. 2017. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544: 59–64. 10.1038/nature21429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stik G, Vidal E, Barrero M, Cuartero S, Vila-Casadesús M, Mendieta-Esteban J, Tian TV, Choi J, Berenguer C, Abad A, et al. 2020. CTCF is dispensable for immune cell transdifferentiation but facilitates an acute inflammatory response. Nat Genet 52: 655–661. 10.1038/s41588-020-0643-0 [DOI] [PubMed] [Google Scholar]
- Su JH, Zheng P, Kinrot SS, Bintu B, Zhuang X. 2020. Genome-scale imaging of the 3D organization and transcriptional activity of chromatin. Cell 182: 1641–1659.e26. 10.1016/j.cell.2020.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemata N, Samson RY, Bell SD. 2019. Physical and functional compartmentalization of archaeal chromosomes. Cell 179: 165–179.e18. 10.1016/j.cell.2019.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares-Cadete F, Norouzi D, Dekker B, Liu Y, Dekker J. 2020. Multi-contact 3C reveals that the human genome during interphase is largely not entangled. Nat Struct Mol Biol 27: 1105–1114. 10.1038/s41594-020-0506-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedeschi A, Wutz G, Huet S, Jaritz M, Wuensche A, Schirghuber E, Davidson IF, Tang W, Cisneros DA, Bhaskara V, et al. 2013. Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature 501: 564–568. 10.1038/nature12471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin BD, González-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. 2012. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 150: 934–947. 10.1016/j.cell.2012.06.051 [DOI] [PubMed] [Google Scholar]
- Tran NT, Laub MT, Le TBK. 2017. SMC progressively aligns chromosomal arms in Caulobacter crescentus but is antagonized by convergent transcription. Cell Rep 20: 2057–2071. 10.1016/j.celrep.2017.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojanowski J, Frank L, Rademacher A, Grigaitis P, Rippe K. 2021. Transcription activation is enhanced by multivalent interactions independent of phase separation. bioRxiv 10.1101/2021.01.27.428421 [DOI] [PubMed] [Google Scholar]
- van Koningsbruggen S, Gierlinski M, Schofield P, Martin D, Barton GJ, Ariyurek Y, den Dunnen JT, Lamond AI. 2010. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell 21: 3735–3748. 10.1091/mbc.e10-06-0508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schaik T, Vos M, Peric-Hupkes D, Hn Celie P, van Steensel B. 2020. Cell cycle dynamics of lamina-associated DNA. EMBO Rep 21: e50636. 10.15252/embr.202050636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Belmont AS. 2017. Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell 169: 780–791. 10.1016/j.cell.2017.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Delrow J, Henikoff S. 2001. Chromatin profiling using targeted DNA adenine methyltransferase. Nat Genet 27: 304–308. 10.1038/85871 [DOI] [PubMed] [Google Scholar]
- Varier RA, Outchkourov NS, de Graaf P, van Schaik FMA, Ensing HJL, Wang F, Higgins JMG, Kops GJPL, Timmers HTM. 2010. A phospho/methyl switch at histone H3 regulates TFIID association with mitotic chromosomes. EMBO J 29: 3967–3978. 10.1038/emboj.2010.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertii A, Ou J, Yu J, Yan A, Pagès H, Liu H, Zhu LJ, Kaufman PD. 2019. Two contrasting classes of nucleolus-associated domains in mouse fibroblast heterochromatin. Genome Res 29: 1235–1249. 10.1101/gr.247072.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vian L, Pękowska A, Rao SSP, Kieffer-Kwon KR, Jung S, Baranello L, Huang SC, El Khattabi L, Dose M, Pruett N, et al. 2018. The energetics and physiological impact of cohesin extrusion. Cell 173: 1165–1178.e20. 10.1016/j.cell.2018.03.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri Rudan M, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, Hadjur S. 2015. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep 10: 1297–1309. 10.1016/j.celrep.2015.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieux-Rochas M, Fabre PJ, Leleu M, Duboule D, Noordermeer D. 2015. Clustering of mammalian Hox genes with other H3K27me3 targets within an active nuclear domain. Proc Natl Acad Sci 112: 4672–4677. 10.1073/pnas.1504783112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Brandão HB, Le TBK, Laub MT, Rudner DZ. 2017. Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science 355: 524–527. 10.1126/science.aai8982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss FD, Calderon L, Wang YF, Georgieva R, Guo Y, Cvetesic N, Kaur M, Dharmalingam G, Krantz ID, Lenhard B, et al. 2021. Neuronal genes deregulated in Cornelia de Lange syndrome respond to removal and re-expression of cohesin. Nat Commun 12: 2919. 10.1038/s41467-021-23141-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wike CL, Guo Y, Tan M, Nakamura R, Shaw DK, Díaz N, Whittaker-Tademy AF, Durand NC, Aiden EL, Vaquerizas JM, et al. 2021. Chromatin architecture transitions from zebrafish sperm through early embryogenesis. Genome Res 31: 981–994. 10.1101/gr.269860.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong X, Hoskins VE, Harr JC, Gordon M, Reddy KL. 2020. Lamin C regulates genome organization after mitosis. bioRxiv 10.1101/2020.07.28.213884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood C, Tonegawa S. 1983. Diversity and joining segments of mouse immunoglobulin heavy chain genes are closely linked and in the same orientation: implications for the joining mechanism. Proc Natl Acad Sci 80: 3030–3034. 10.1073/pnas.80.10.3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz G, Várnai C, Nagasaka K, Cisneros DA, Stocsits RR, Tang W, Schoenfelder S, Jessberger G, Muhar M, Hossain MJ, et al. 2017. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J 36: 3573–3599. 10.15252/embj.201798004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Emerson DJ, Gilgenast TG, Titus KR, Lan Y, Huang P, Zhang D, Wang H, Keller CA, Giardine B, et al. 2019. Chromatin structure dynamics during the mitosis-to-G1 phase transition. Nature 576: 158–162. 10.1038/s41586-019-1778-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang Y, Chen Y, Gholamalamdari O, Wang Y, Ma J, Belmont AS. 2021. TSA-seq reveals a largely conserved genome organization relative to nuclear speckles with small position changes tightly correlated with gene expression changes. Genome Res 31: 251–264. 10.1101/gr.266239.120 [DOI] [PMC free article] [PubMed] [Google Scholar]