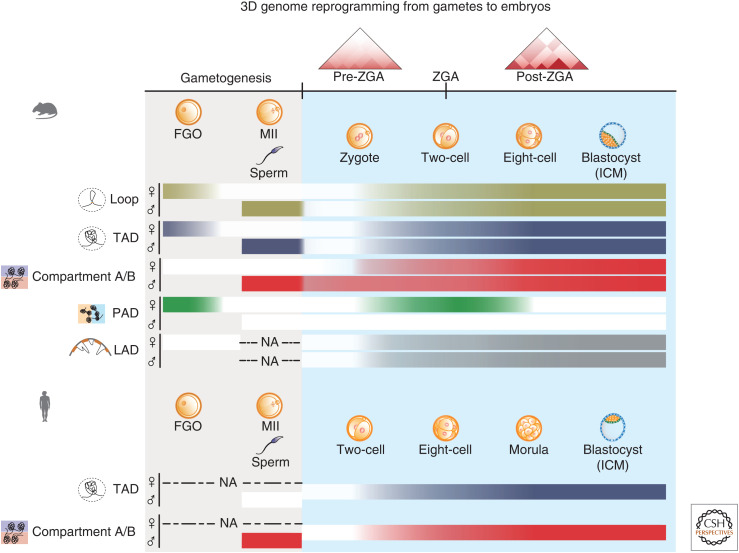
Figure 3.
3D genome dynamics in gametes and early embryos. The schematics showing the dynamics of higher-order chromatin structure from gametes to embryos in mouse and human. In mice, full-grown oocytes (FGOs) exhibit cohesin-independent, H3K27me3-marked compartmental domains (Polycomb-associating domains [PADs]). By contrast, topologically associating domains (TADs) become weak and compartments A/B are undetectable in FGOs. Lamina-associated domains (LADs) are also undetected in FGOs. In MII oocytes, chromatin adopts a mitotic-like chromatin structure, where compartments A/B, PADs, TADs, and loops are all lost. By contrast, mouse mature sperm shows canonical A/B compartmentalization, TADs, and loops. After fertilization, chromatin structure is dramatically dispersed, which shows weakened compartments, TADs, and loops. The genome structure is then gradually reestablished during preimplantation development. PADs briefly reappear specifically on the maternal allele from one-cell to eight-cell stages. LADs (gray) are de novo established after fertilization. Human embryos also exhibit diminished higher-order structure after fertilization, followed by consolidation of TADs and compartments A/B after zygotic genome activation (ZGA). Notably, human sperm has no typical TADs possibly due to the lack of CTCF protein. The strength of TADs, compartmentalization (compartments A/B and PADs/iPADs), and LADs are indicated by the shades of the bars. (NA) Data not available, (ICM) inner cell mass.