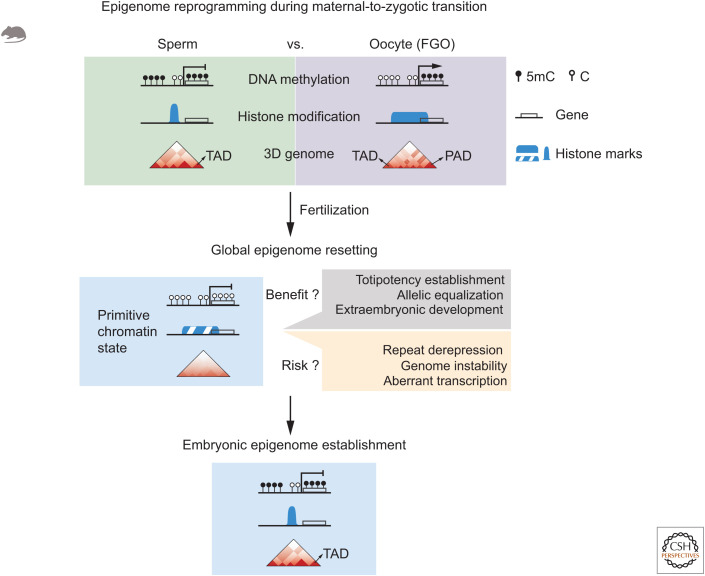
Figure 4.
Epigenome reprogramming during maternal-to-zygotic transition (MZT). Mouse oocytes, but not sperm, adopt noncanonical epigenomes including DNA hypomethylated nontranscribing regions, broad domains of key histone modifications, and unique 3D genome structures. After fertilization, the embryos undergo global epigenome resetting, featured by the global loss of repressive epigenetic marks (such as DNA methylation), widespread presence of active histone marks, and global chromatin relaxation. The global resetting might be beneficial for the establishment of totipotency, allelic epigenome equalization, and extraembryonic tissue development. Meanwhile, it may also expose embryos for risks of repeat derepression, genome instability, and aberrant transcription. After zygotic genome activation (ZGA), the embryos stepwise establish embryonic epigenomes to restore DNA methylation, histone marks, and higher-order chromatin structure. (FGO) Full-grown oocyte, (TAD) topologically associating domain, (PAD) Polycomb-associating domain.