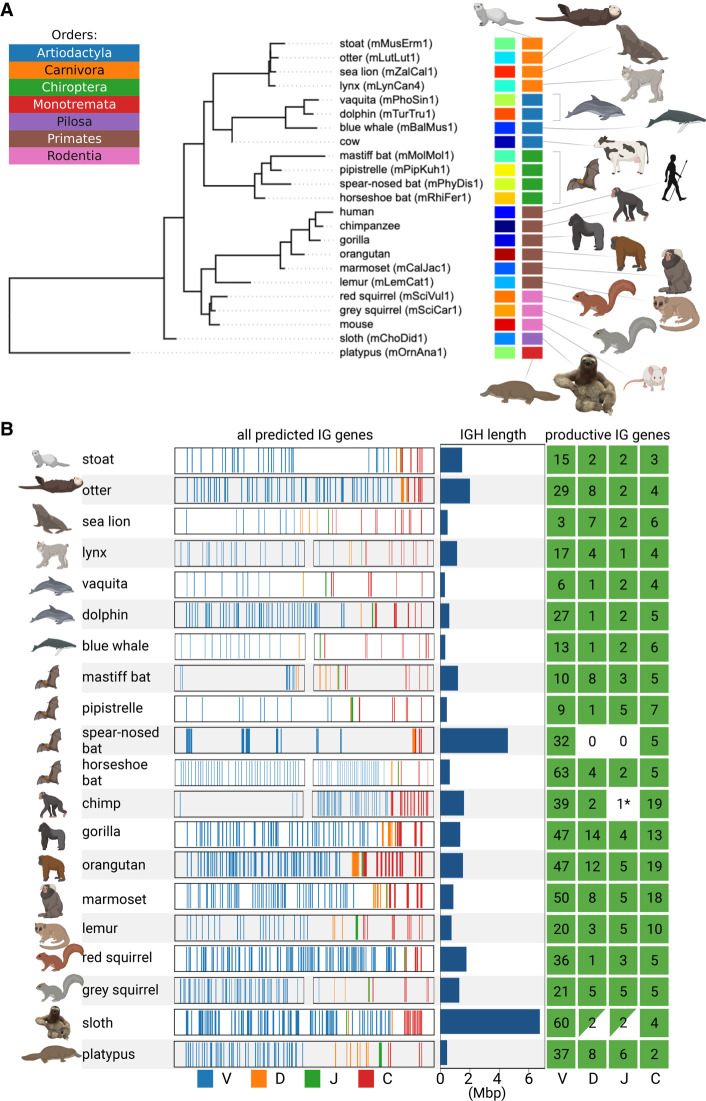
Figure 2.
Information about the IGH loci in 20 target mammalian species. (A) The phylogenetic tree formed by 20 target and three reference species. The tree was subsampled from the Tree of Life constructed by Hedges et al. (2015). Each species is shown by its common name and a VGP identifier specified in parentheses if available. Each species is encoded by a unique color (left vertical color panel) and a color representing its order (right vertical color panel). The list of orders is shown in the upper left corner. Here and below visualization was performed using the BioRender and Iroki (Moore et al. 2020) tools. (B) Information about the IGH loci of 20 target species. Each line corresponds to a target species and shows fragments of the IGH locus with positions of candidate V (blue), D (orange), J (green), and C (red) genes. For six out of 20 species, the IGH-end covers <80% of the predicted IGH locus length (lynx, blue whale, mastiff bat, horseshoe bat, chimpanzee, and grey squirrel). We showed both the IGH-start and the IGH-end for these six species and only the IGH-end for the remaining species. For a better visualization, all IGH loci are shown as having the same length that does not reflect their real lengths (the bar plots next to the IGH loci show the predicted lengths). The map on the right shows the counts of the productive V, D, J, and C genes identified in the IGH locus. A C gene is classified as productive if its translated regions (defined by the closest human C gene) does not contain stop codons. Nonzero counts are shown in green. The green triangles indicate partially found and (likely) partially missing D and J genes in the sloth IGH locus. Although IterativeIGDetective did not identify any J genes in two species (chimpanzee and spear-nosed bat) within the boundaries of the IGH loci, it found a highly conserved candidate J gene in a short contig in the chimpanzee assembly (denoted as 1* on the map of the right). IterativeIGDetective did not identify any candidate J genes in the spear-nosed bat, presumably because all its RSSJs did not pass the likelihood threshold.