Abstract
Accurate classification of pulmonary nodules in the CT images is critical for early detection of lung cancer as well as the assessment of the effect from COVID-19. In this paper, we propose a computer-aided classification method for lung nodules using expert knowledge. We use a decoupling metric learning model to describe the deep characteristics of the nodules and then calculate the similarity between the current nodule and the nodules in the database. By analyzing the returned nodules with the diagnosis information, we obtain the expert knowledge of similar nodules, based on which we make the decision of the current nodule. The proposed method has been evaluated on the benchmark LIDC-IDRI dataset and achieved an accuracy of 95.7% and AUC of 0.9901. The proposed classification method can have a variety of applications in lung cancer detection, diagnosis and therapy.
Keywords: Lung nodule, classification, CT, convolutional neural networks (CNN), expert knowledge
1. INTRODUCTION
Almost one-quarter of all cancer deaths are due to lung cancer 1. In addition, the pandemic of coronavirus disease 2019 (COVID-19) has infected one hundred million people in almost 200 countries 2,3. It is found that most lung cancers showed small nodules at the baseline screening 4. And the nodules are included in the chest computed tomography (CT) of the patients with COVID-19 pneumonia 5. Early diagnosis of pulmonary nodules can provide an opportunity for designing effective treatment and making care plans. Classification of lung nodules has many clinical applications. CT is a viable diagnostic tool for the detection of lung cancer and COVID-19. It can provide valuable information to identify lung nodules. Due to the enormous amount of CT data and complexity of the nodules, observer performance studies are time-consuming. A computer-aided classification of lung nodules can accomplish a quantitative evaluation fast.
The computer-aided classification methods for lung nodules mainly employ the machine learning methods, especially deep learning methods recently. Wang et al proposed an adaptive-boost deep learning to train a strong classifier for invasiveness classification of subsolid nodules in chest CT images, using multiple 3D convolutional neural network (CNN)-based weak classifiers. It used the prior-feature learning to reduce the computations by sharing the CNN layers between all weak classifiers 6. Lin et al presented an interpretable end-to-end computer-aided detection and diagnosis tool for pulmonary nodules on CT using deep learning-based methods. It used a hierarchical semantic convolutional neural network to classify whether a nodule was benign or malignant and generate semantic features 7. Liu et al proposed a multi-task deep model with margin ranking loss network for automated lung nodule analysis. It explored the relatedness between lung nodule classification and attribute score regression with multi-task deep learning model. And it employed the siamese network in a weight-shared manner with a margin ranking loss to model the nodule heterogeneity and encourage the discrimination capability on ambiguous nodule cases 8. Xie et al proposed a semi-supervised adversarial classification model which used the both labeled and unlabeled data for the lung nodule classification. It consisted of an adversarial autoencoder-based unsupervised reconstruction network, a supervised classification network, and learnable transition layers that enable the adaption of the image representation ability learned by unsupervised network to supervised network. It employed three semi-supervised adversarial classification models for the multi-view knowledge-based collaborative learning 9. Hussein et al presented a 3D CNN based graph regularized sparse multi-task learning for the malignancy determination of lung nodules 10. Xie et al proposed an algorithm for lung nodule classification that fused the texture, shape, and deep model-learned information at the decision level. It employed a gray level co-occurrence matrix-based texture descriptor, a Fourier shape descriptor to characterize the heterogeneity of nodules, and a deep convolutional neural network to automatically learn the feature representation of nodules on a slice-by-slice basis 11. Dey et al proposed four two-pathway CNNs, including a basic 3D CNN, a novel multi-output network, a 3D DenseNet, and an augmented 3D DenseNet with multi-outputs 12 . Xie et al proposed a multi-view knowledge-based collaborative deep model to separate malignant from benign nodules. The model learned 3D lung nodule characteristics by decomposing a 3D nodule into nine fixed views and joint the nine a knowledge-based collaborative submodel for each view for classifying the lung nodules with an adaptive weighting scheme learned during the error back propagation 13.
Those methods focus on extracting and analyzing the CT image features for the classification of lung nodules. In fact, radiologists interpret the CT scans for distinguishing malignant from benign nodules, based on not only CT imaging information, but also the historical clinical experience. They use the accumulated clinical experience to assist the diagnosis of nodules. Inspired by that, we propose a computer-aided classification of lung nodules with the aid of the clinical expert knowledge. We retrieve the similar nodules from the dataset as the expert knowledge for aiding the classification of the nodules to improve the diagnostic accuracy.
2. METHOD
The proposed lung nodules classification method with the expert knowledge contains two parts, which are the lung nodule characteristics generation and the expert knowledge based benign-malignant classification. The radiologists always make a diagnosis on a lung nodule according to its appearance and their clinical knowledge. By imitating the doctor’s diagnosis behavior, we extract the deep features of the lung nodules to represent the characteristic, and search the similar nodules in the database to acquire the clinical knowledge. Fig. 1 shows the overview of the proposed classification method.
Fig 1.
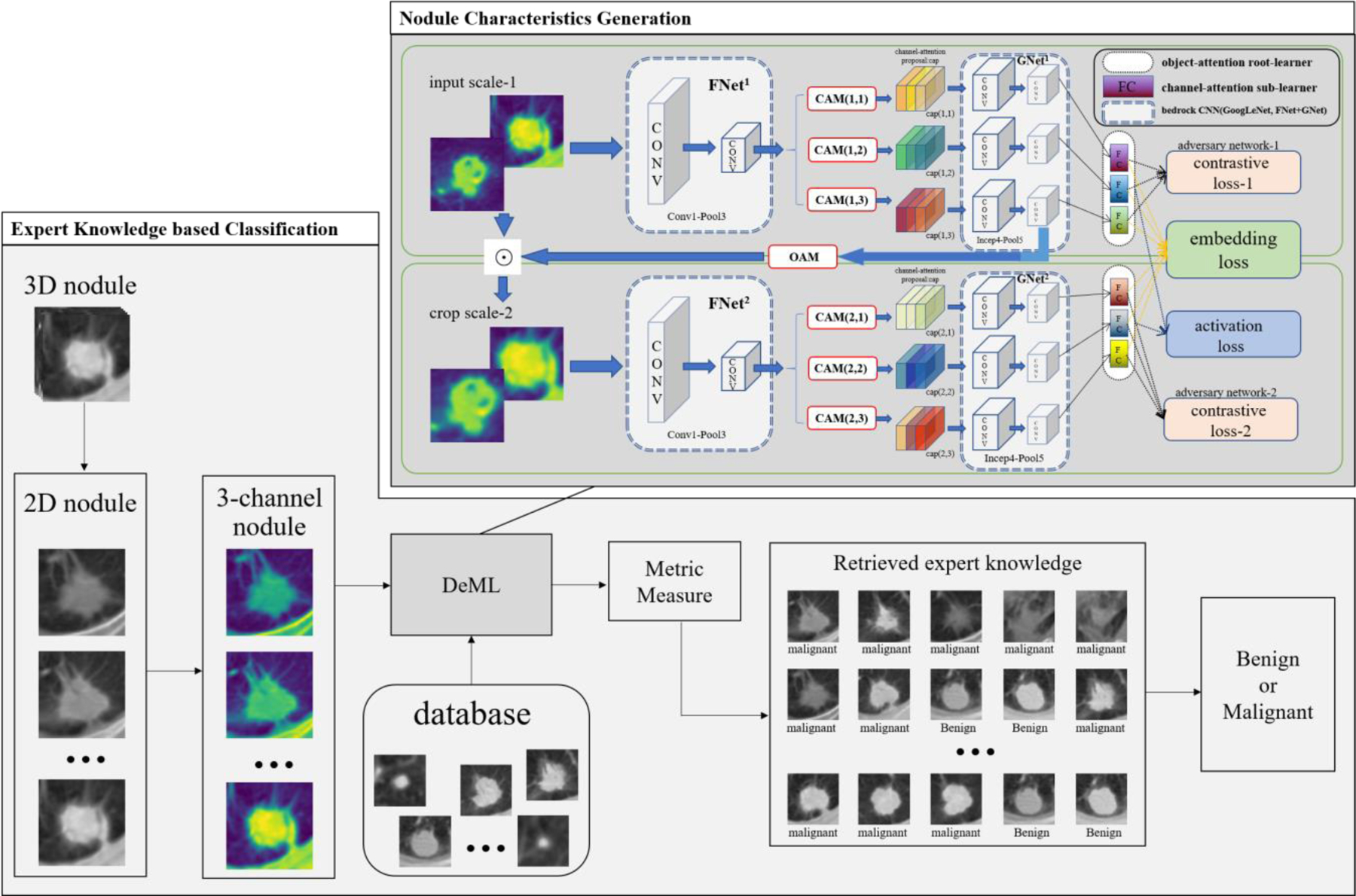
Overview of the proposed expert knowledge-based classification method.
2.1. Nodule characteristics generation
In the feature extraction stage, we use the Decoupling Metric Learning (DeML) model 14 for describing the characteristics of the lung nodule. It can improve the ability of model recognition and generalization by decoupling the input lung nodule characteristics into multiple tasks. The model has four parts: the GoogLeNet, the object-attention module (OA), the channel-attention module (CAM), and the adversary network.
In the dark gray part of the Fig. 1, we can see that we change the CT image of lung nodule into the image with three channels, and have two OA root learners and three sub-learners per root learner to obtain rich information. The Conv1 to Pool5 of GoogLeNet to build the CNN infrastructure, where the Conv1-Pool3 make up the FNet and Incept4-Pool5 make up the GNet. Then the OA module part crops the nodule images into an appropriate scale for focusing on the import information. The CAM part trains the different sub-learner for focusing the different attention part of the nodules. To obtain the diversity enhance of sub-learners, the adversary network is used to minimize the difference between CA learners. In addition, we involve the attribution information of the lung nodules, including the lobulation, calcification and speculation, for the comprehensive representation of the lung nodules.
2.2. Expert knowledge based benign/malignant classification
In the classification stage, we represent a three-dimensional pulmonary nodule by several two-dimensional slices. We select M 2D slices and conduct the M retrieval processing. For each 2D nodule, we calculate the cosine similarity of its feature and the features of the nodules in the database, and return the most similar N 2D database nodules. The N nodules with the diagnosis information are considered as the expert knowledge to guide the classification of the current nodule. If more than half of the M☓N diagnosis results of the returned nodules are benign (malignant), we classified the current pulmonary nodule into benign (malignant). The decision can be made by:
| (1) |
where C is the final diagnosis decision, max(a, b) is a function returning the bigger value of the integer a and b, abs(x) is the absolute value of x, Di,j is the diagnosis result of the j-th returned nodule in the database for the i-th retrieval processing, and M and N are the number of slices of the 3D nodule and returned images in the database.
2.3. Evaluation metrics
We use the accuracy and area under curve (AUC) to evaluate the classification performance. The two evaluation metrics are defined as follow:
| (2) |
where NTP, NFP, NTN, and NFN denote the number of true positives, false positives, true negatives, and false negatives, respectively, NP and NN are the numbers of positive samples and negative samples, and RANKi is the rank of the i-th positive sample.
3. EXPERIMENTS AND RESULTS
3.1. Database
The classification method of lung nodule was evaluated on the LIDC-IDRI dataset 15, which is the largest publicly available lung nodule dataset. We extracted 1530 nodules and 9646 slices. We use the three signs, lobulation, calcification and speculation, for assisting the benign/malignant classification. We selected 80% and 20% nodules as training set and testing set, respectively, and make sure that the nodules in the training set and testing set come from different patients for avoiding the bias in measuring performance.
We implemented the proposed method based on the Caffe framework with one Nvidia Tesla V100 16G GPU. In the process of pre-processing, we normalize the spatial resolution of each image to 1.0 mm, and resize all the images to a uniform size of 56 ☓ 56. In the learning process, we use Adam optimizer with learning rate 1e-5 and weight decay 2e-4. The batch size is 16 and the training iteration is 20k, for our experiments.
3.2. Classification accuracy with different returned numbers N and slice numbers M
We combine M 2D slices to form a whole 3D nodule, and we search the similar top N images as the expert knowledge for the classification of the current nodule. We test the classification accuracy with the different returned numbers, N, and the different numbers of slices, M, as shown in Fig. 2. In Fig. 2, we can see that our classification method can obtain a highest accuracy of 96.1% when M is four and N is fifteen. Although M and N are bigger, that means we can get more expert knowledge, the classification accuracy don’t increase. A possible reason is that too many slices may create some interference information. On the whole, we can obtain the average accuracy of 94.61% with the standard deviation of 1%, which indicates that our method is not only effective, but also robust.
Fig. 2.

The classification accuracy with different returned numbers N and slice numbers M.
3.3. Classification performance
Since most nodules across five slices, and we find that we can obtain the stable results when N is bigger than seven in Fig. 2, we define M to be five and return top seven similar slices as the expert knowledge for a more robust evaluation. We can obtain the accuracy of 95.7% and AUC with 0.9901. The receiver operating characteristic (ROC) curve of the classification method is as shown in Fig. 3.
Fig. 3.
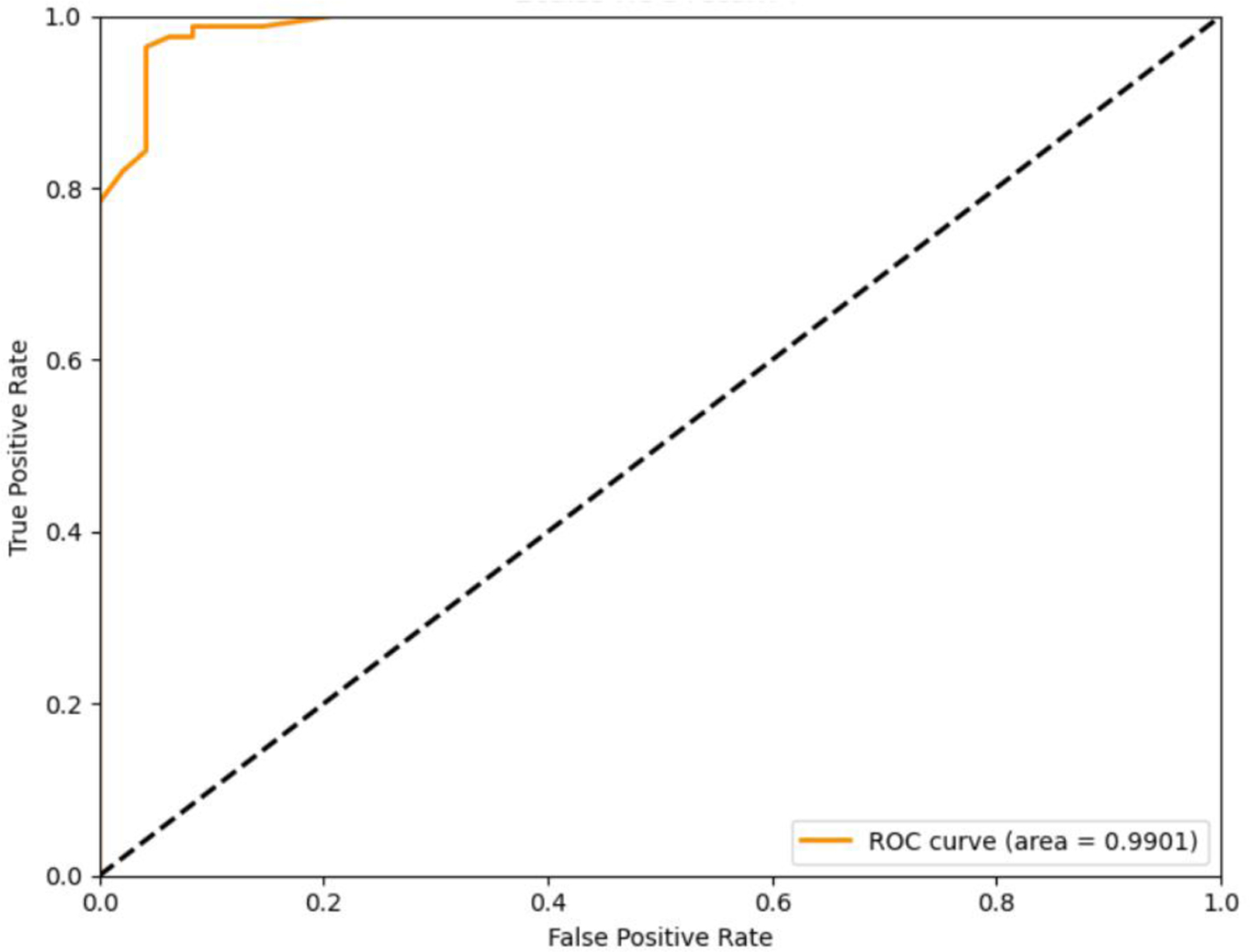
The ROC curve of the classification method.
3.4. Comparison results
We compare our method with the other classification methods of lung nodule, and the compared results are shown in Table 1. In Table 1, we can see that our method can obtain the highest classification accuracy and biggest AUC, and we can obtain the increase rates of 2.3% for accuracy and 1.1% for AUC compared with the other methods. That proves our method is effective and outstanding.
Table 1.
The Compared Results
4. CONCLUSION
In this paper, an expert knowledge-based method is proposed for the classification of the lung nodule in CT images. We use the DeML to extract the deep features of the nodules and search the similar nodules. The clinical diagnosis of retrieved similar nodules is taken as expert knowledge for making the decision and improving the classification accuracy of lung nodules. The experimental results show that our method obtained satisfactory classification performance and can be suitable for various applications in lung disease diagnosis.
ACKNOWLEDGMENTS
This research was supported in part by the Natural Science Foundation of China (No. 61901234), China Postdoctoral Science Foundation (2018M641635), and the Beijing Municipal Science and Technology Project under Grant (Z181100001918002).
REFERENCES
- [1].Siegel RL, Miller KD and Jemal A, “Cancer statistics, 2020,” CA: A Cancer Journal for Clinicians, 70(1): 7–30 (2020). [DOI] [PubMed] [Google Scholar]
- [2].Callaway E “Time to use the p-word? Coronavirus enter dangerous new phase,” Nature, 579: 12 (2020). [DOI] [PubMed] [Google Scholar]
- [3].Johns Hopkins University & Medicine, COVID-19 resource, https://coronavirus.jhu.edu/map.html
- [4].Henschke CI, Yankelevitz DF, Mirtcheva R, et al. , “CT screening for lung cancer: frequency and significance of part-solid and nonsolid nodules,” American Journal of Roentgenology, 178(5): 1053–1057 (2002). [DOI] [PubMed] [Google Scholar]
- [5].Shi H, Han X, Jiang N, et al. , “Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study, “ The Lancet Infectious Diseases, 20(4), 425–434 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wang J, Chen X, Lu H, et al. , “Feature‐shared adaptive‐boost deep learning for invasiveness classification of pulmonary subsolid nodules in CT images, “ Medical Physics, 47(4):1738–49 (2020). [DOI] [PubMed] [Google Scholar]
- [7].Lin Y, Wei L, Han SX, et al. , “EDICNet: An end-to-end detection and interpretable malignancy classification network for pulmonary nodules in computed tomography,” Proc. SPIE on Medical Imaging, 11314, 113141H (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liu L, Dou Q, Chen H, et al. , “Multi-task deep model with margin ranking loss for lung nodule analysis,” IEEE Transactions on Medical Imaging, 39(3):718–28 (2020). [DOI] [PubMed] [Google Scholar]
- [9].Xie Y, Zhang J, Xia Y. “Semi-supervised adversarial model for benign–malignant lung nodule classification on chest CT,” Medical Image Analysis, 57:237–48 (2019). [DOI] [PubMed] [Google Scholar]
- [10].Hussein S, Kandel P, Bolan CW, et al. , “Lung and pancreatic tumor characterization in the deep learning era: novel supervised and unsupervised learning approaches,” IEEE Transactions on Medical Imaging, 38(8):1777–87 (2019). [DOI] [PubMed] [Google Scholar]
- [11].Xie Y, Zhang J, Xia Y, et al. , “Fusing texture, shape and deep model-learned information at decision level for automated classification of lung nodules on chest CT”. Information Fusion, 42: 102–110 (2018). [Google Scholar]
- [12].Dey R, Lu Z, Hong Y. “Diagnostic classification of lung nodules using 3D neural networks”. IEEE 15th International Symposium on Biomedical Imaging (ISBI), 774–778 (2018). [Google Scholar]
- [13].Xie Y, Xia Y, Zhang J, et al. , “Knowledge-based Collaborative Deep Learning for Benign-Malignant Lung Nodule Classification on Chest CT”. IEEE Transactions on Medical Imaging, 38(4):991–1004 (2018). [DOI] [PubMed] [Google Scholar]
- [14].Chen B, Deng W. “Hybrid-Attention based Decoupled Metric Learning for Zero-Shot Image Retrieval,” Proceeding of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2750–2759 (2019). [Google Scholar]
- [15].Armato SG 3rd, McLennan G, Bidaut L, et al. “The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A completed reference database of lung nodules on CT scans”. Medical Physics, 38: 915–931 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]


