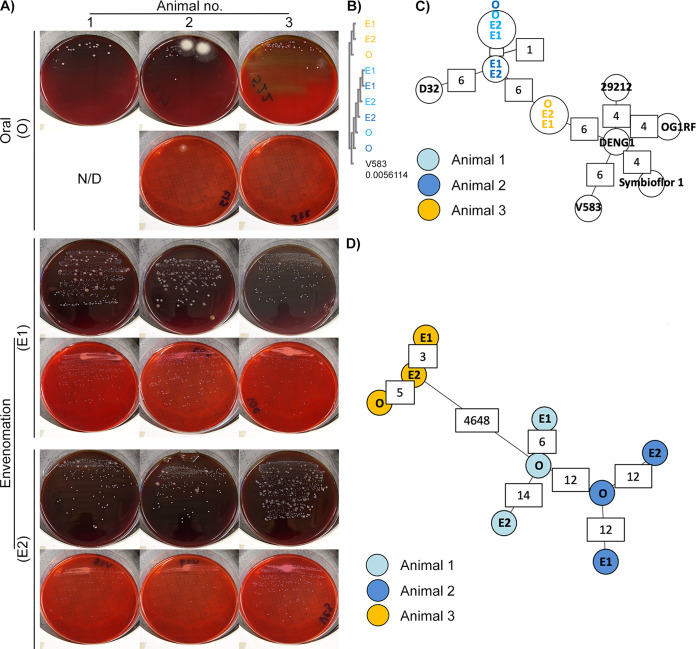
FIG 3.
Whole-genome sequencing identifies viable bacteria in N. nigricollis venom as two animal-specific E. faecalis strains. (A) White punctate colonies were recovered in blood agar (upper panels) and MacConkey agar (lower panels) blind cultures of individual oral swab (O) and two consecutive envenomation samples (E1 and E2) obtained from three captivity N. nigricollis snakes. N/D, none detected. (B) Blind multiple-sequence alignment (ClustalO followed by ClustalW phylogeny) of homologous sequences across the de novo assembled genomes against the E. faecalis V583 katA gene (distance to V583 katA indicated in the V583 track) suggests two sequence groups reflecting the history and housing of the host animals. (C) Blind MST construction based on the MLST of the N. nigricollis-derived isolates against nine E. faecalis reference genomes again separates samples into two distinct clusters that reflect the history and housing of the host animals. Partially available allele data are included in this analysis, and instances of allelic differences between nearest neighbors are annotated in white boxes. (D) Blind complete genome MLST against a custom schema generated using three closely related E. faecalis reference genomes clusters these isolates by animal of origin (animal 1, light blue; animal 2, dark blue; animal 3, orange). The host animal color scheme depicted in panel D is also used in panels B and C.