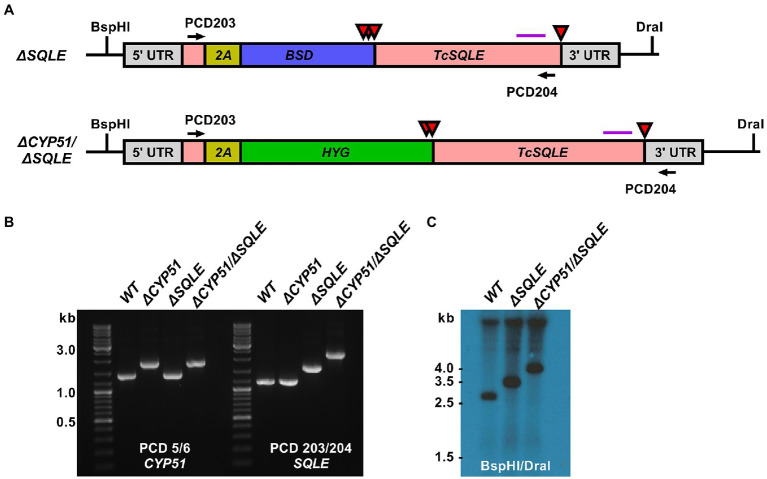
Figure 5.
(A) Predicted TcSQLE loci following homology-directed repair template integration shown. The TcSQLE coding region is shown in pink, skip peptide in yellow. Blasticidin-S deaminase (blue) was used in WT parasites while hygromycin B phosphotransferase (green) was used for transfection of ΔCYP51 parasites already carrying the blasticidin-S deaminase gene. Red triangles indicate the positions of stop codons. The purple horizontal line corresponds to the predicted binding location for the Southern blot probe; BspHI and DraI sites in the genome are indicated. (B) Primers flanking TcSQLE were used to amplify the locus and identify clones with increased band size on an agarose gel. (C) Southern blot following digestion of genomic DNA with BspHI/DraI and hybridization with a probe binding to the 3′ region of TcSQLE.