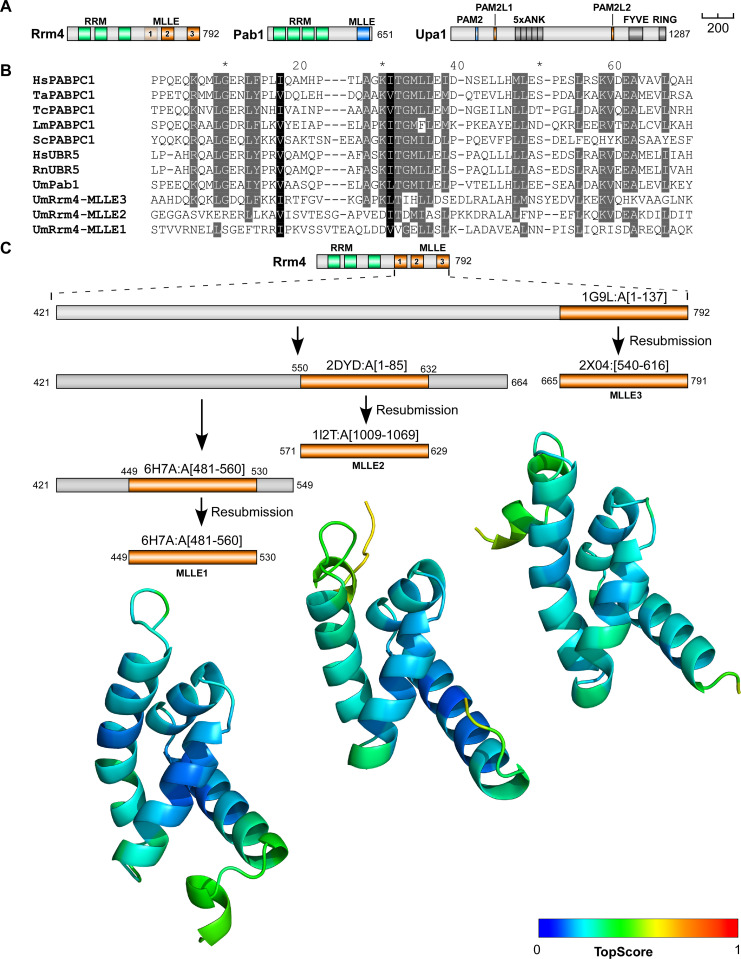
Fig 1. The C-terminal half of Rrm4 contains three MLLE domains.
(A) Schematic representation of protein variants drawn to scale (bar, 200 amino acids, number of amino acids indicated next to protein bars) using the following colouring: dark green, RNA recognition motif (RRM); orange, MLLERrm4 domains; dark blue, MLLEPab1; light blue PAM2; light orange PAM2L sequence (PL1–2) Ankyrin repeats (5xANK), FYVE domain, and RING domain of Upa1 are given in dark grey. (B) Sequence alignment of previously determined MLLE domains showing the degree of similarity to the three Rrm4-MLLE domains and the positions (Hs—Homo sapiens, Ta—Triticum aestivum, La—Leishmania major, Sc—Saccharomyces cerevisiae, Tc—Trypanosoma cruzi, Rn—Rattus norvegicus, Um—Ustilago maydis, PABPC1, Pab1 –poly [A]-binding protein, UBR5—E3 ubiquitin-protein ligase). Accession number and sequence coverage are listed in S1 Table. Multiple sequence alignment was performed by ClustalW. (C) Identification and modelling of C-terminal MLLE domains of Rrm4. The iterative process is depicted graphically. The best-identified template for each run, and the region of that template that aligns with Rrm4, are displayed (see also S1A Fig for the templates used for the final models). The structural models obtained are shown for the span of the first identified template and are coloured according to their per-residue TopScore, where the scale from 0 to 1 indicates a low to high local structural error.