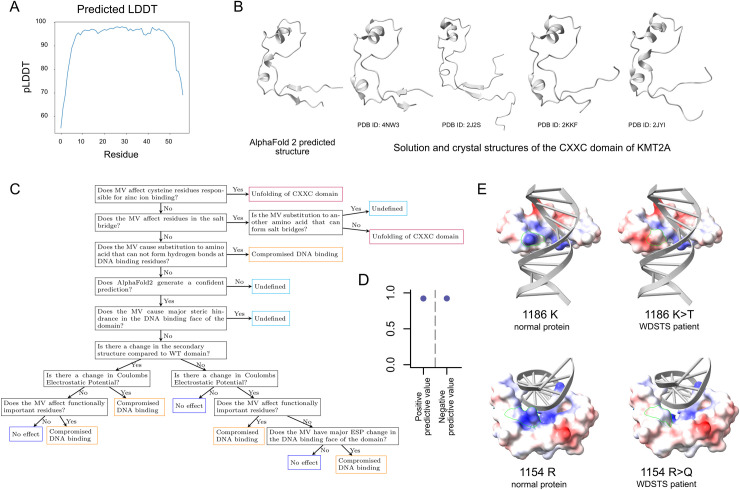
Fig 3. An AlphaFold2-based variant effect classification scheme for the CXXC domain of KMT2A.
(A) Predicted LDDT values for the CXXC domain of KMT2A. (B) The AlphaFold2-predicted and experimentally determined structures of the CXXC domain of KMT2A. (C) The variant effect classification scheme. See Methods for details on the derivation of the scheme. (D) The positive and negative predictive value that the classifier shown in (C) attained on a hold-out test set consisting of 41 missense variants (see main text and Methods for details). (E) The position of two different missense variants in the 3D structure of the CXXC domain, in conjunction with a 3D representation of the engaged DNA backbone. For comparison, the same representation is shown for the normal protein as well (PBD* ID: 4NW3). The surface of the domain is color-coded based on the electrostatic potential, with red indicating a negative charge and blue a positive charge. Both variants lead to a decreased electrostatic potential in a residue important for DNA binding.