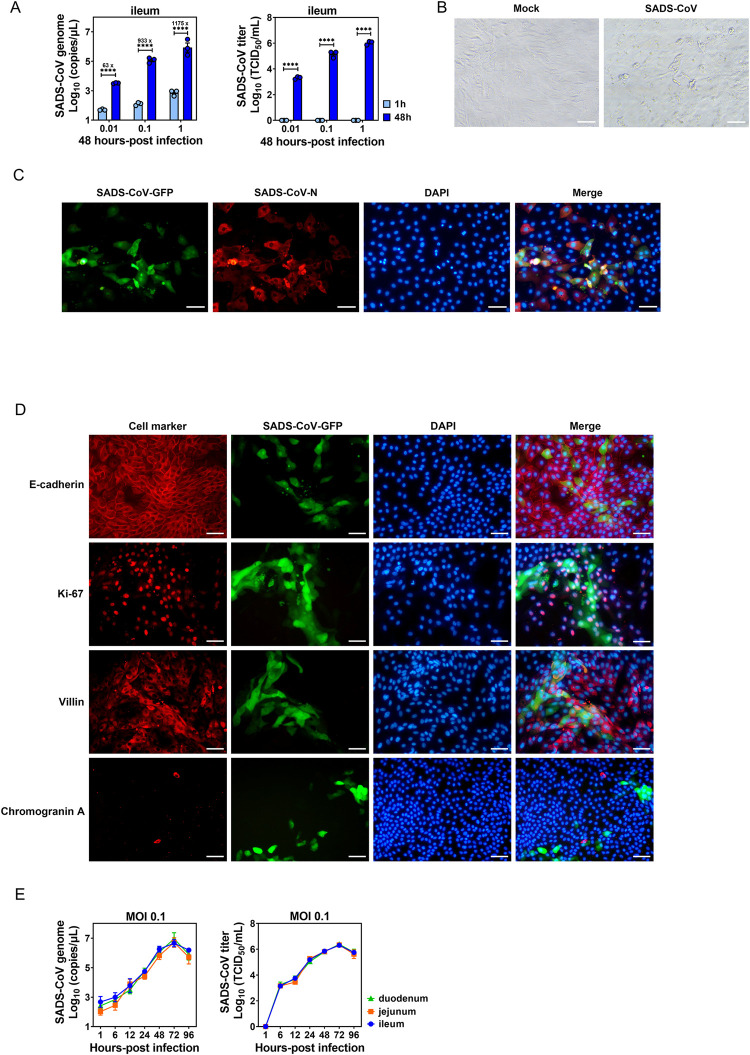
Fig 2. Porcine intestinal enteroid (PIE) cultures support SADS-CoV replication.
(A) Ileal PIE monolayers were incubated with medium or SADS-CoV-GFP at the indicated MOIs on a rotary shaker for 1 h at 37°C. The incubated monolayers were washed three times with PBS and harvested at 1 or 48 hpi. Supernatant RNA was extracted and the number of SADS-CoV-GFP genome copies was determined by RT-qPCR and viral titer was determined by a standard TCID50 assay. (B) Cytopathic effect (CPE) was observed in ileal PIEs infected with SADS-CoV at 48 hpi (scale bar, 50 μm). (C) Immunofluorescence assay (IFA) was performed using rabbit anti-SADS-CoV-N polyclonal antibody and Alexa Fluor 594-conjugated anti-rabbit IgG as secondary antibody, and nuclei (blue) were visualized by DAPI (scale bar, 50 μm). (D) Colocalization of SADS-CoV-GFP and cellular markers (red) including E-cadherin (epithelial tight junction), Ki-67 (proliferating cells), villin (enterocytes) and chromogranin A (enteroendocrine cells), and nuclei (blue) stained by DAPI (scale bar, 50 μm). (E) Duodenal, jejunal and ileal PIE monolayers were incubated with SADS-CoV-GFP at MOI = 0.1 and the SADS-CoV-GFP replication kinetic was determined by RT-qPCR or TCID50. Data are from three independent experiments. P values were determined by unpaired two-tailed Student’s t test. *: p < .05; **: p < .01; ***: p < .001; ****: p < .0001; ns, not significant.