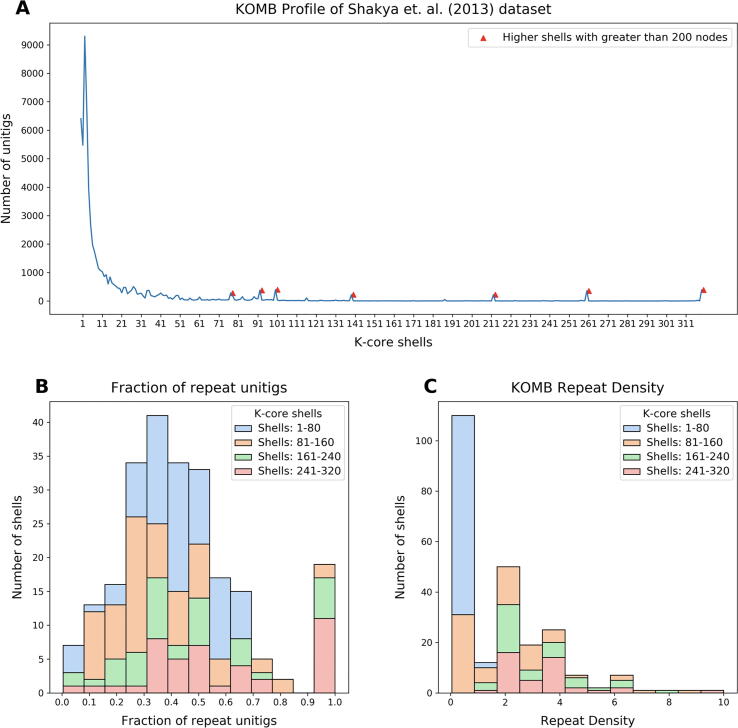
Fig. 6.
Characterization of a synthetic metagenome sample using KOMB. A. KOMB profile of the Shakya et al. (2013) dataset representing the shell number on the x-axis and the number of unitigs in the y-axis. Red triangles indicate higher shells with greater than 200 nodes, which represent clique or clique-like regions in the hybrid unitig graph. B. Histogram representing the fraction of unitigs in each of the shells that are repeats as determined by comparing with the nucmer output. C. KOMB repeat density is defined as the average copy number per number of genomes for the repeat unitigs in the shell (see Eq. 2). Larger shells have repeats with high copy number and more specific to a single (or group of related) organisms. For figures B. and C., shell 0 (disconnected nodes) and shells that contained no unitigs are not considered. Shells are split into four different groups (1–80, 81–160, 160–240, 241–320) for visualization.