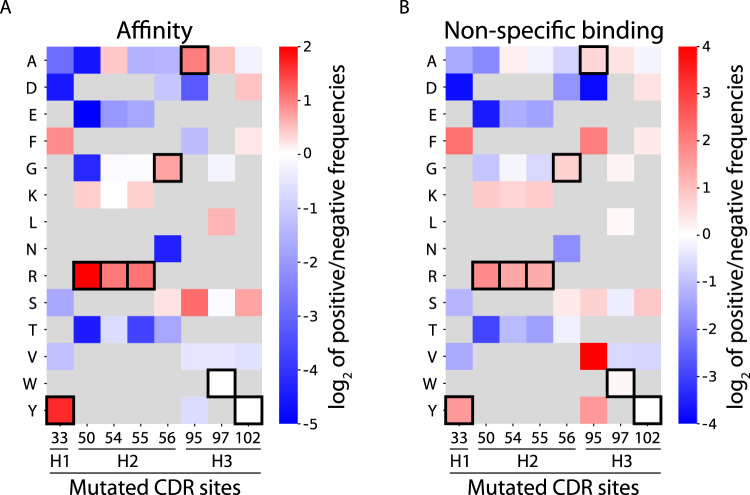
Fig. 2. Levels of CDR residue enrichment in the sorted emibetuzumab libraries are similar for high affinity and high non-specific binding selections.
Calculated ratios between the amino acid frequencies at each mutated site for library samples selected for (A) high affinity relative to low affinity and (B) high non-specific binding (ovalbumin and soluble membrane proteins) relative to low non-specific binding. Red mutations appeared more frequently in the high affinity and high non-specific binding library samples, while blue mutations appeared more frequently in the low affinity and low non-specific binding library samples. Wild-type residues are highlighted with black boxes, and residues that were not sampled in the libraries are indicated with gray shading.