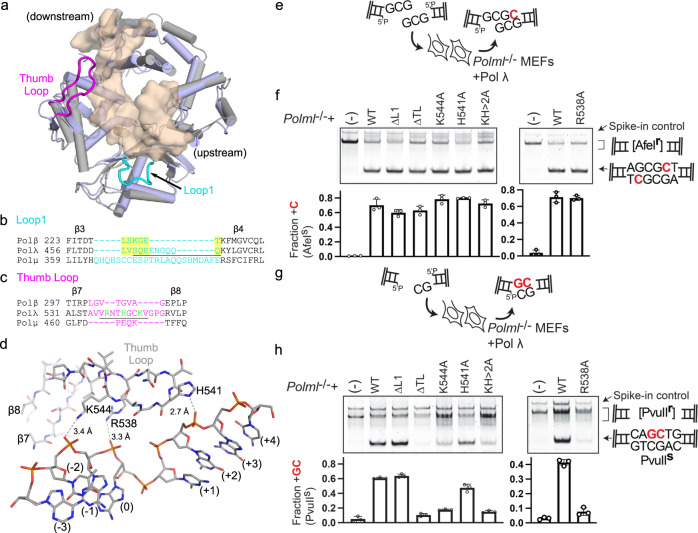
Fig. 3. Determinants of DSB substrate specificity by Polλ.
a Structural superposition of 1nt SSB crystal structures of Polλ (PDB ID code 1XSN11, protein in gray and DNA in transparent khaki surface) and Polμ (PDB ID code 4M0412, protein in light blue). Loop1 and the ‘thumb loop’ regions of Polλ are shown in cyan and magenta, respectively. b Structure-based sequence alignment of Loop1 (cyan) in Pols β, λ, and μ (structurally-equivalent regions highlighted in yellow). The Polλ ΔL1 variant was generated by deleting Ser463-Gln471 (underlined) and replacing them with Lys462-Thr465 from Polβ13. c Sequence alignment of ‘thumb loop’ (TL, magenta) of Pols β, λ and μ. The Polλ ΔTL variant was generated by deleting Val537-Val545 of Polλ (underlined) and fusing the ends of β-strands 7 and 8 with a single glycine residue. Thumb loop mutations used in this study are marked in green. d Interactions of the TL with an SSB substrate (PDB ID code 1XSN11, gray). Hydrogen bonds with the DNA template strand are shown as black dashed lines. e DNA fragments with partly complementary ends (GCG3ʹ overhangs) were introduced into Polm−/−Poll−/− cells (Polml−/−). f Representative gel electrophoresis (top) identification of the fraction of NHEJ that is AfeI-sensitive and polymerase-dependent. Quantification of the average fraction of polymerase-dependent repair using GCG3ʹ DSB substrates from triplicate electroporations (bottom; error bars represent the standard deviation). g As in (e), except using noncomplementary blunt and GC3ʹ overhangs. h As in (f), except using PvuII sensitivity to identify polymerase-dependent repair.