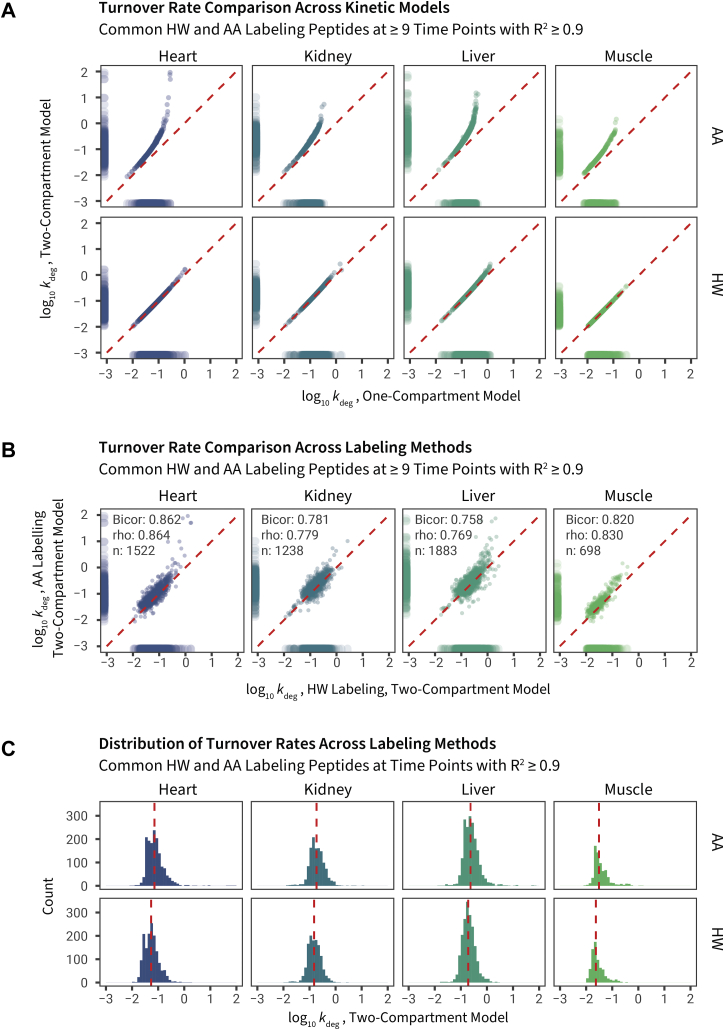
Fig. 6.
Comparisons of turnover rate constants across labels and kinetic models. A, degradation rate constants (kdeg) were obtained for peptides from four tissues using amino acid (AA) or heavy water (HW) labeling and were fitted using a one-compartment (x-axis) or two-compartment (y-axis) kinetic model to derive the first order kdeg (plotted on a log base 10 scale). Data points are peptide time series commonly quantified in HW and AA experiments containing 1 lysine and with ≥9 time points and fitted with R2 ≥ 0.9. Red dashed line: unity. Marginal distribution shows data density. B, scatterplots of shared peptides quantified by HW and AA in each tissue using the two-compartment model (quantified time points ≥9, R2 ≥ 0.9, 1 lysine). Numbers denote robust correlation (biweight midcorrelation; bicor) coefficients, Spearman correlation coefficient (rho), and number of comparisons (n). Each data point is one peptide-charge time series. C, histogram showing distribution of kdeg across tissues and labels. Red dashed lines denote medians.