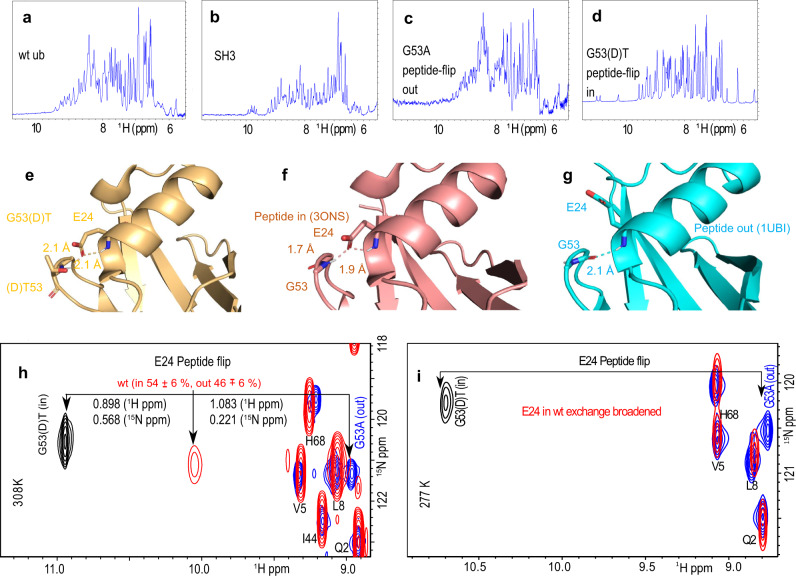
Fig. 5. NMR spectroscopy characterization of SH3c and ubiquitin wt and mutants G53(D)T and G53A.
a–d 1H NMR spectra of the amide region showing for all proteins a range of chemical shifts up to or more than 9 ppm, indicating that the two mutants of ubiquitin as well as the wt and the construct of SH3c are well-folded in the buffer condition used for the experiments. e–g crystal structures of the G53(D)T mutant as well as the two crystal structures of wt ubiquitin showing the “in” conformation of the peptide bond (f) and the “out” conformation (g). The G53(D)T (e) mutant of ubiquitin (golden ribbon) has a similar poise of the G53 peptide-bond and the side-chain of E24 as the wild-type ubiquitin in the “in” conformation (f, magenta ribbon, PDB: 3ONS). The side-chain of the (D)T53 is shown in stick representation. The dihedral angles for (D)T53 (ϕ : 109.7°; ψ : 18.6°) show that the molecules is locked into the peptide-flip “in” conformation. For comparison, the dihedral angles of G53 in the “in” conformation are ϕ : 98.6°, and ψ : −25.6° (PDB: 3ONS) and “out” conformation along the peptide-flip mode, are ϕ : −82.9°, and ψ : −8.9° (PDB: 1UBI). HSQC spectra of the wt, the G53(D)T and G53A mutant of ubiquitin at 308K (h) and 277 K (i). The black arrows indicate the positions of the NH resonance of E24. The G53(D)T and G53A mutants were designed to redistribute the populations to “in” and “out” conformations along the peptide-flip mode, respectively.