Figure 2.
cNMF algorithm reveals distinct expression signatures
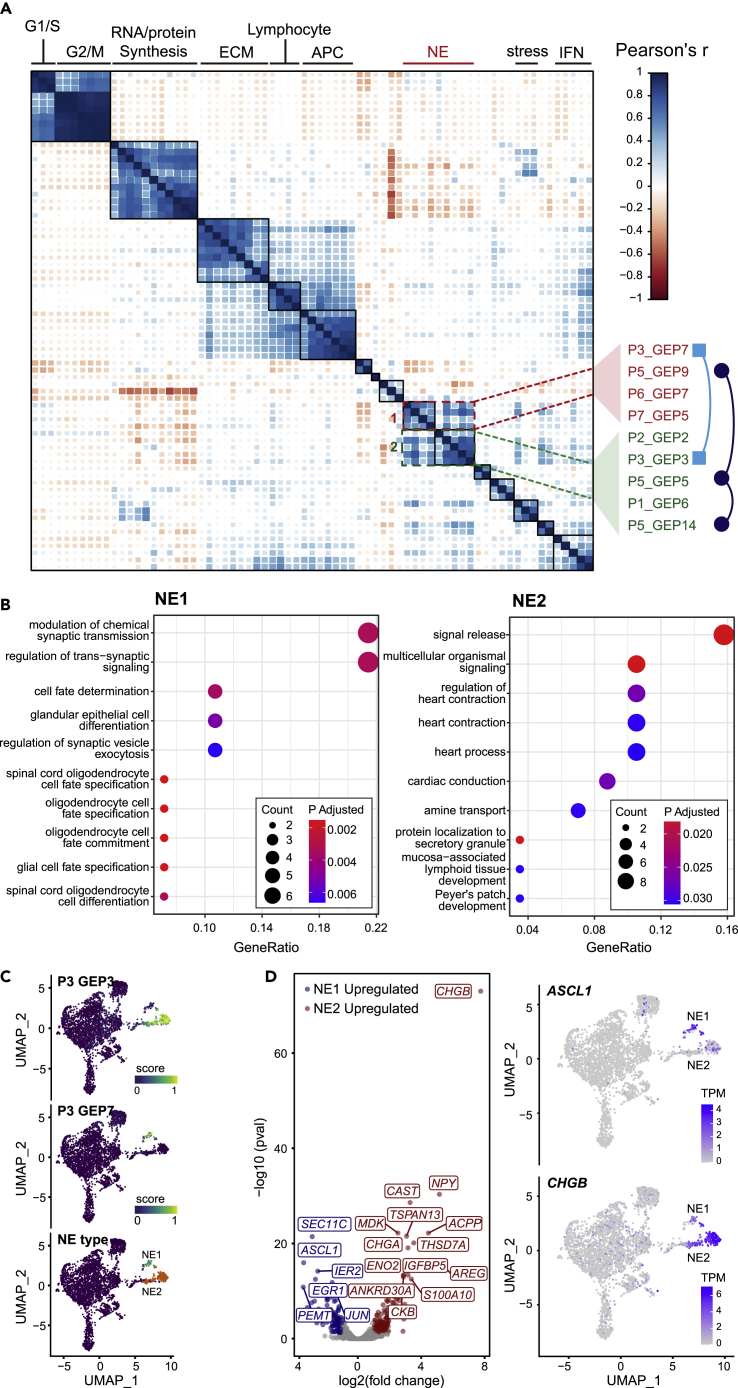
(A) Pairwise correlation clustering of intra-tumoral GEPs. 71 GEPs were derived by cNMF from seven tumors and formed ten consensus modules, whose biological significance was predicted by GO analysis (top). GEPs in two NE modules were annotated (right) (also see Table S6).
(B) GO analysis of genes over-represented in NE1 and NE2, respectively (hypergeometric distribution test adjusted by FDR <5%) (also see Figure S3, Table S7).
(C) UMAP plot of malignant cells from P3. Cells are colored by their NE subtype (right) and the min-max normalized activity score of the corresponding GEPs (left, middle).
(D) Volcano plot of the differentially expressed genes (DE-Gs) between NE1 and NE2 cells of P3 (Wilcoxon rank-sum test adjusted by FDR <5%). Blue, upregulated in NE1; Red, upregulated in NE2; gray, | log 2-fold change <1 | or adjusted P-value > 0.05. Expression of marker genes of note, ASCL1 and CHGB, for NE1 and NE2, respectively, were shown in the UMAP plot. TPM, edtranscript per million (normalized value). (also see Figure S4, Table S8).