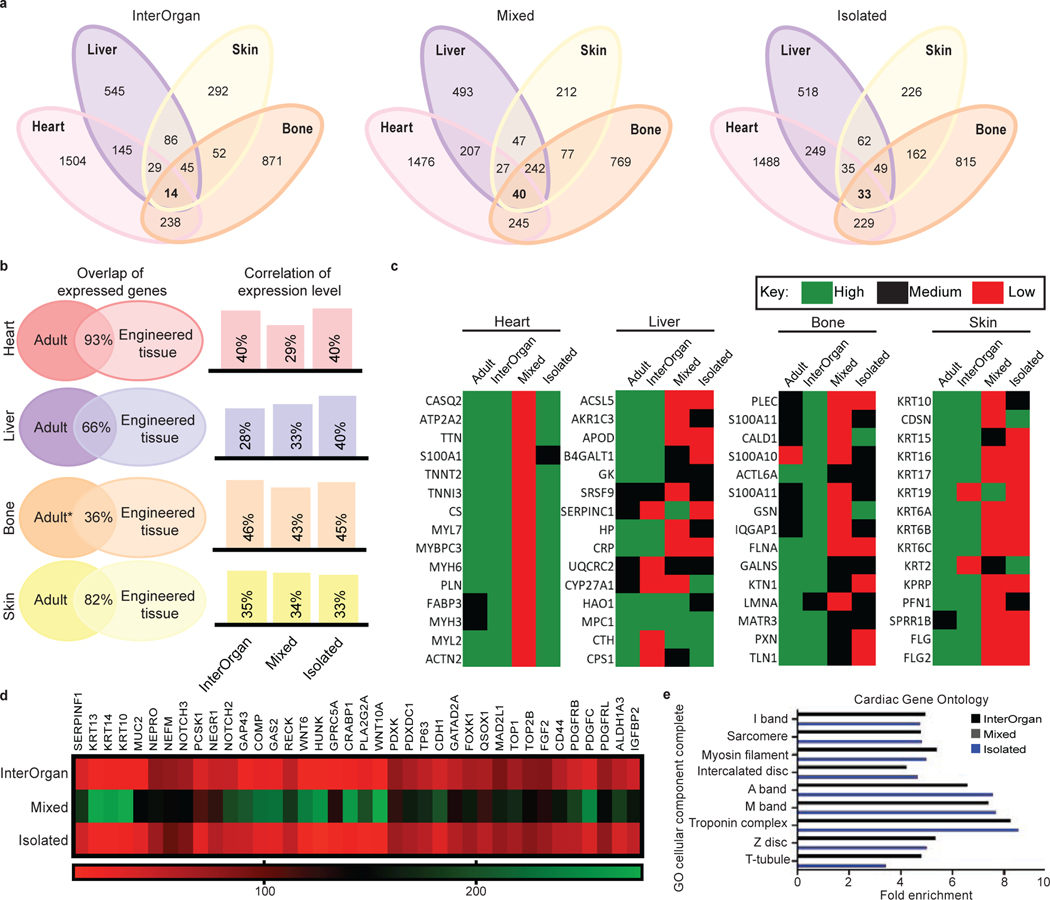
Figure 5 |. Proteomic analysis confirms biological fidelity of InterOrgan tissue chip in comparison to gold standards and adult organs.
a, When comparing all engineered organs within each experimental group, GO analysis identified gene pathways shared amongst the different organs. b, Comparison between published adult data and engineered tissues demonstrates high similarities in the shared expression of genes to native tissue (left); additional comparisons in how the expression level of these shared genes within each experimental group correlates to adult tissue is presented as well (right). c, Within each organ system, top proteins of interest were identified via the Human Protein Atlas and compared to adult tissue. d, As cardiac tissues were of highest biological fidelity, we identified a number of candidate proteins that are often found in off-target tissues, including in development of epithelial, osteochondral, and neural tissues, for example. e, GO analysis identified cardiac-specific, adult-like structural components present in the InterOrgan (dark blue) and Isolated (light blue) groups, but not at all in the Mixed condition (gray).