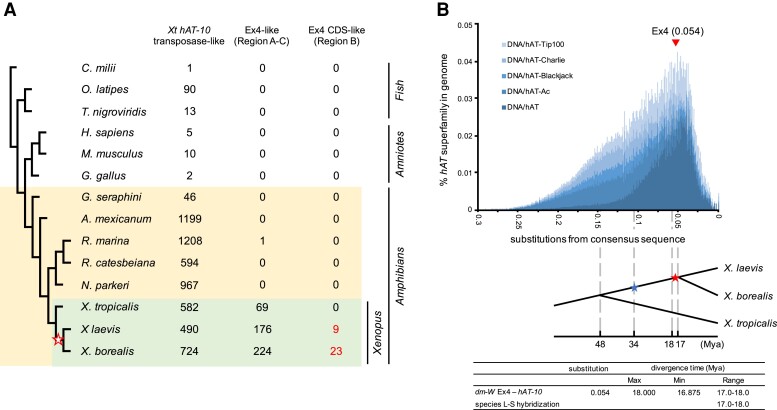
Fig. 2.
Molecular evolution of hAT superfamily, hAT-10 family, and hAT-10-derived Ex4 in Xenopus frogs. (A) Copy numbers of Xt hAT-10 transposase (Tp)-like sequences, hAT-10-derived Ex4-like sequences (A–C in fig. 1B), hAT-10-derived Ex4-CDS-like sequences [B in A] in 14 vertebrate species, including anuran amphibians (Xenopus laevis, X. borealis, X. tropicalis, Nanorana parkeri, Rana catesbeiana, and Rhinella marina), an urodele amphibian (Ambystoma mexicanum), a caecilian amphibian (Geotrypetes seraphini), a bird (Gallus gallus), mammals (Homo sapiens and Mus musculus), teleost fish (Oryzias latipes and Tetraodon nigroviridis), and a cartilaginous fish (Callorhinchus milii). GenBank assembly accession of 11 species except for the three Xenopus species used is shown in supplementary fig. S3, Supplementary Material online. (B) A repeat landscape of hAT superfamily consisting of the five families, as inferred in the X. laevis genome using RepeatMasker (upper). The y-axis and x-axis show percentages of each family on the genome and Jukes-Cantor-corrected divergence, respectively. The estimated divergence time of the hAT-10-derived regions on and around dm-W Ex4 from the hAT-10 consensus sequence is shown by a triangle on the landscape. After the ancestors of X. laevis and X. tropicalis diverged at 48 Ma, speciation and hybridization of the predicted L and S species occurred at 34 and 17–18 Ma, respectively (Session et al. 2016).