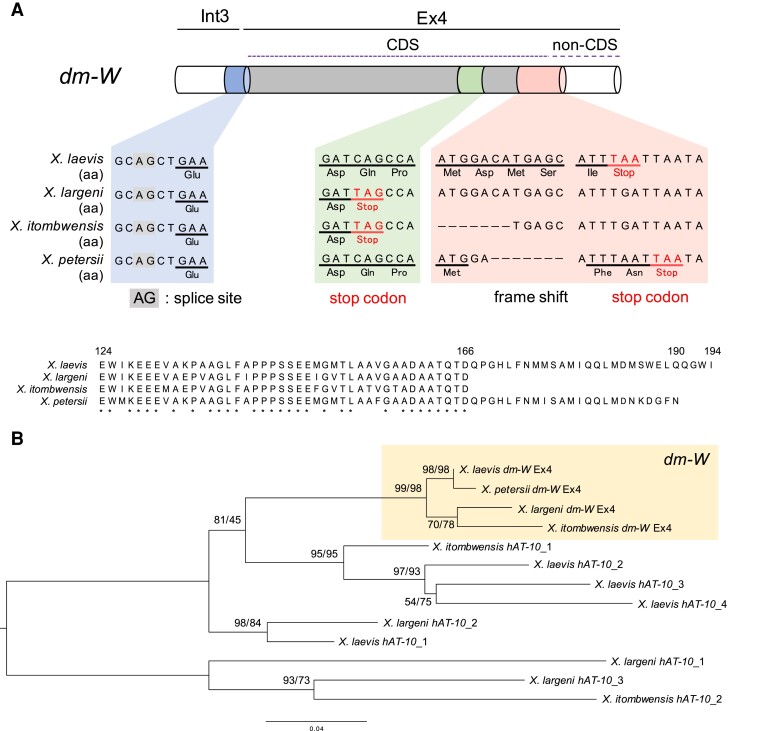
Fig. 3.
The nucleotide and deduced amino acid sequence alignments of the Ex4-CDS of dm-W among four allopolyploid Xenopus species. (A) A multiple nucleotide sequence alignment within and adjacent to Ex4-CDS of X. laevis, X. largeni (MCZ-A cryogenic 333), X. itombwensis (MCZ-A A136197), and X. petersii. Red font highlights the positions of TAG and TAA stop codons. (B) A multiple alignment of the Ex4-encoded amino acid sequences from the four DNA sequences in (A) (upper) and the ML phylogenetic tree of the Ex4 sequences and/or their corresponding hAT-10-derived sequences from X. laevis, X. largeni, X. itombwensis, and X. petersii (lower). Numbers at each node denote the ML/NJ bootstrap percentages of 1000 replicates.