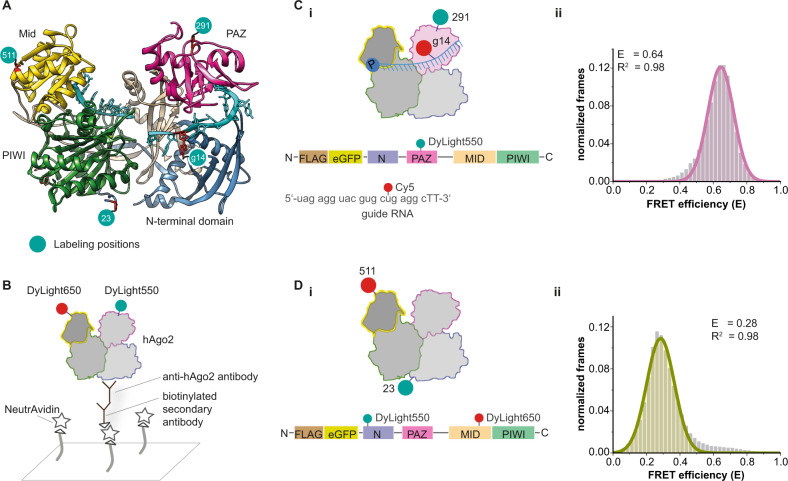
Fig. 1. Single-molecule FRET measurements using site-specifically labeled human Ago2.
A Crystal structure of a hAgo2-guide RNA complex (pdb 4W5N). Green dots indicate labeling positions. The amino acids (F23, H291, T511) were mutated to p-azido-L-phenylalanine (AzF) to enable site-specific labeling. B Immobilization strategy to allow single-molecule FRET measurements using TIRF microscopy. C (i) Labeling positions for intermolecular FRET measurements using DyLightTM550-labeled hAgo2PAZ and a guide RNA14Cy5. (ii) FRET efficiency histogram for the setup shown in (i). D (i) Labeling positions of hAgo2N/Mid for intramolecular FRET measurements. (ii) FRET efficiency histogram for hAgo2N/Mid. Both histograms show data of at least three independent measurements. Data were fitted to a single Gaussian equation. Source data provided as a Source Data file.