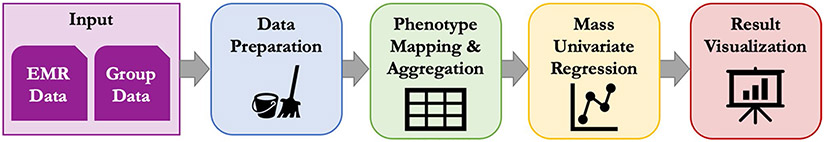
Fig. 2.
PheDAS analysis pipeline. Inputs to the pipeline include EMR data (ICD-9, ICD-10, or CPT codes) and group data (disease group, sex, race, etc.). The data is first prepared for analysis via case–control matching and censoring. Next, the EMR data is mapped to a set of predefined phenotypes (PheWAS or ProWAS Codes) and aggregated across each subject’s record. Mass univariate regression is then performed across all phenotypes, where a target variable is modeled as a function of the phenotype plus any relevant covariates (such as sex or race) to determine the relationship between the target variable and each phenotype. Finally, the results are visualized to facilitate interpretation of target variable-phenotype relationship significance and effect size