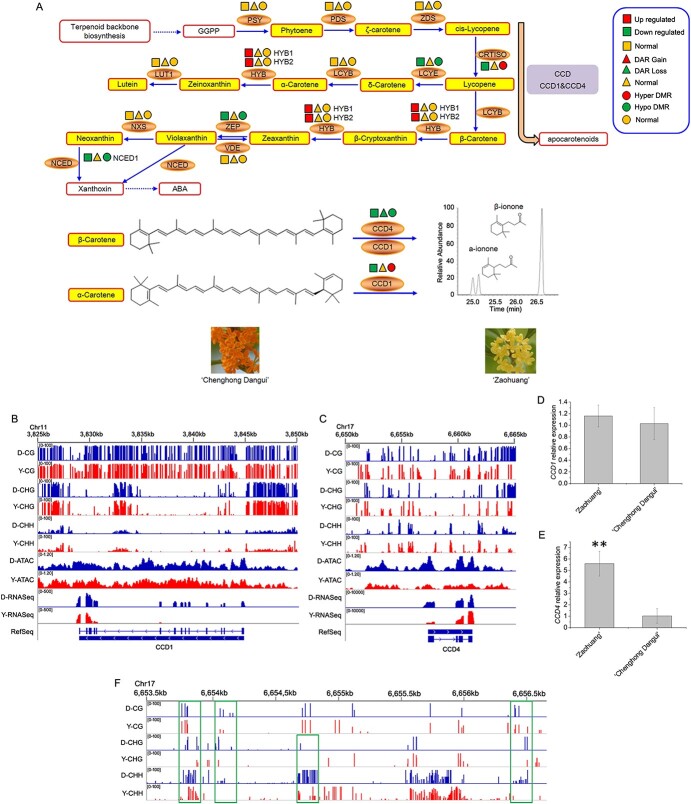
Figure 6.
Effects of methylation level and chromatin accessibility on the expression of genes related to ionone production in sweet osmanthus ‘Chenghong Dangui’ petals (‘Zaohuang’-vs-‘Chenghong Dangui’). A, Methylation level and chromatin accessibility analysis of related genes in ionone production. GGPP: geranylgeranyl diphosphate, PSY: phytoene synthase, PDS: phytoene desaturase, ZDS: ζ-carotene desaturase, CRTISO: carotene isomerase, LCYB: lycopene β-cyclase, LCYE: lycopene ε-cyclase, HYB: β-carotene hydroxylase, LUT1: carotene ε-monooxygenase, ZEP: zeaxanthin epoxidase, VDE: violaxanthin de-epoxidase, NXS: neoxanthin synthase, NCED1: 9-cis-epoxycarotenoid dioxygenase, ABA: abscisic acid, CCD1: carotenoid cleavage dioxygenase 1, CCD4: carotenoid cleavage dioxygenase 4. B–C, DNA methylation level and chromatin accessibility analysis of CCD1 and CCD4. D–E, qRT-PCR analysis of CCD1 and CCD4 in petals of the two cultivars at the full flowering stage. F, DNA methylation level 3 kb upstream of CCD4. Higher methylation level regions 3 kb upstream of CCD4 in ‘Chenghong Dangui’ are marked with green boxes. Y, ‘Zaohuang’; D, ‘Chenghong Dangui’.